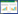中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- Zhenhong Gao, Zhiwei Zhang, Xiaoguang Xu, Jinxin Che, Yan Zhang, Yanlin Liu, Junling Shi. 2018
- 高振红, 张志伟, 徐晓光, 车金鑫, 张艳, 刘延琳, 师俊玲. 2018
- Genomic analysis reveals the biosynthesis pathways of diverse secondary metabolites and pinoresinol and its glycoside derivatives in Phomopsis sp.XP-8
- 基因组分析揭示拟茎点霉XP-8产松脂醇及其糖苷等多种次级代谢产物的合成途径
- Acta Microbiologica Sinica, 58(5): 939-954
- 微生物学报, 58(5): 939-954
-
文章历史
- 收稿日期:2017-12-13
- 修回日期:2018-01-19
- 网络出版日期:2018-02-07
2. College of Food Science and Engineering, Qingdao Agriculture University, Qingdao 266109, Shandong Province, China;
3. Key Laboratory for Space Bioscience and Biotechnology, School of Life Sciences, Northwestern Polytechnic University, Xi'an 710072, Shaanxi Province, China;
4. College of Enology, Northwest A & F University, Yangling 712100, Shaanxi Province, China
2. 青岛农业大学食品科学与工程学院, 山东 青岛 266109;
3. 西北工业大学生命学院, 空间生物重点实验室, 陕西 西安 710072;
4. 西北农林科技大学葡萄酒学院, 陕西 杨凌 712100
Lignan is one of the major functional compounds that are mainly found in plants[1]. Pinoresinol (pin) is a simple lignan in nature and has been found to possess multiple functions, such as anti-cancer[2], anti-tumor[3], antifungal[4] and anti-inflammatory[5] activities, as well as prevention of hormone-dependent diseases, such as cardiovascular diseases, hyperlipidemia and breast cancer[6-8]. However, the natural production of lignans, including pin is very low in nature. The identification of lignan biosynthesis pathway and key genes would provide essential information for large-scale production of lignan compounds using microbial fermentation via genetically modified microorganisms. The lignan biosynthesis pathway has been well known as the phenylpropanoid pathway that is widely found in plants[9], but not currently found in microorganisms[10-11].
In plants, the phenylpropanoid pathway plays a central role for the biosynthesis of a diverse group of compounds, such as lignin, coumarins, stilbenes, flavonoids, isoflavonoids, and lignans[12]. In this pathway (Figure 1), D-Erythrose-4P (E4P) from glycolysis and phosphoenolpyruvate (PEP) from pentose phosphate pathway are conversed to chorismate via the shikimate pathway, and then produce phenylalanine, which is transformed into an activated cinnamic acid derivative via the actions of phenylalanine ammonia lyase (PAL), trans-cinnamate 4-monooxygenase (CYP73A), 4-coumarate-CoA ligase (4CL), and specific branch pathways[13-14].

|
| Figure 1 The central role of phenylpropanoid pathway in biosynthesis of secondary metaboliates in plants. |
Many endophytic fungi from plants have been found to possess the capability to produce plant derived compounds that have anti-tumor, anti-inflammatory, antibacterial, and antifungal activities[15-16]. In recent years, endophytic fungi have attracted great attention of scientists due to their potential application in medicine, agriculture, and other industries. Lignans and their glycosides are the major effective components of Eucommia ulmoide Oliv. (E. lmoides)[17]. To date, 32 lignans, including pin, pinoresinol-4-O-β-D- glucopyranoside (PMG), and pinoresinol diglucoside (PDG), have been identified and isolated from E. ulmoides[18]. Several approaches have also been developed to produce pin and its derivatives by chemical or enzymatic methods[19-21]. Phomopsis sp. XP-8, an endophytic fungus isolated from the bark of Tu-Chung (E. ulmoides), was found to produce pin and its monoglucoside (PMG) and diglucoside (PDG)[22], similar as the products found in E. ulmoides[23], providing a potential method to produce pin and its glucoside derivatives by microbial fermentation. Zhang[24] identified that Phomopsis sp. XP-8 could accumulate the intermediate compounds (cinnamic acid and p-coumaric acid) and had key enzymatic activities (PAL, CYP73A, 4CL) in the phenylpropanoid pathway. However, an understanding of pin and its glucoside derivatives biosynthesis at the genetic level should facilitate greater pin and its glucoside derivatives production[25].
In order to identify the phenylpropanoid pathway and pin and its glucosides biosynthesis pathway in Phomopsis sp. XP-8 at the gene level, and to help identify more significant genes as well. De novo genome sequencing, feature analysis, were explored in this study.
1 Materials and methods 1.1 Fungal strains and genomic DNA preparationPhomopsis sp. XP-8, an endophytic fungus previously isolated from the bark of Tu-Chung (Eucommia ulmoides Oliv.) and maintained at the China Center for Type Culture Collection, (Wuhan, China) (code: CCTCC M209291), was selected for genomic analyses in the study. After Phomopsis sp. XP-8 cultivation for 120 h under 28 ℃ and 180 r/min in PDB (containing 200 g potato, 20 g dextrose, and 1000 mL tap water), highly purified fungal DNA extraction was resulted in the construction of libraries using the cetyltrimethyl ammoniumbromide (CTAB) method[26]. DNA quality and quantity were evaluated using standard 0.8% agarose gel electrophoresis, and a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA).
1.2 Genome Sequencing and AssemblyAfter whole genome DNA extraction, DNA sequencing libraries with 600 bp and 3 kb inserts were constructed using Nextera Mate Pair Library Preparation Kit (Illumina, San Diego, CA, USA), following the standard Illumina protocol. The generated 4418637781 reads were quality control-filtered; that is, low-quality reads were removed. High-quality reads were obtained and then assembled by the de novo method using Velvet v1.2.10[27]. Final assembly was evaluated for completeness using CEGMA v2.5[28] and BUSCO v2.0[29], and the resulting assembled sequences were used for gene model prediction, functional annotation, and downstream information analyses. The whole genome sequencing of Phomopsis sp. XP-8 was performed using the Illumina Hiseq 2500 with 96.8-fold coverage. The sequenced reads were assembled into 330 scaffolds (≥1000 bp).
1.3 Gene Prediction and Functional AnnotationThe genes in the Phomopsis sp. XP-8 genome were predicted using various software. Repetitive sequences were de novo predicted using LTR FINDER[30], MITEHunter[31], RepeatScout[32] and PILERDF[33], and then classified according to PASTEClassifier[34]. RepeatMasker[35]was subsequently used against the Repbase[36] TE library and the de novo repeat library to annotate repeats in the Phomopsis sp. XP-8 genome. miRNAs were predicted by BLASTN of genomic sequence slices against miRBase sequences. tRNAs were annotated using tRNAscan-SEv1.3.1[37]. rRNA was annotated by aligning the genomic sequence against the Rfam database by using BLASTn with an e-value ≤1e-5. Protein-coding genes were predicted using Augustus (v3.1 http://bioinf.uni-greifswald.de/augustus/). The predicted CDSs were searched by Gapped BLAST and PSI-BLAST[38] against the NCBI non-redundant protein (Nr) database, Pfam and Swiss-Prot database, Gene Ontology (GO)[39], Cluster of Orthologous Groups[40](COGs), Kyoto Encyclopedia of Genes and Genomes (KEGG)[41]metabolic pathways. The e-value thresholds were all set at ≤1e-5. The antibiotics and secondary metabolite analysis shell (antiSMASH) automatic pipeline (http://antismash.secondarymetabolites.org/) was used to predict gene clusters and the core structure of secondary metabolites in Phomopsis sp. XP-8.
1.4 Gene Family AnalysesOrthologous gene clusters of Phomopsis sp. XP-8 and other reference fungal strains were determined using OrthoMCL v1.4[42] with a default MCL inflation index of 1.5. The result of the OrthoMCL analysis was used to plot Venn diagrams. Protein alignments, of each single-copy ortholog family, were performed using MUSCLE[43]. All the poorly aligned regions were removed. The maximum likelihood phylogenetic tree of the six species included in this analysis was created based using phyML v3.0[44]with bootstrapping for 1000 replicates.
1.5 Analysis of Pinoresinol and its Glycosides Biosynthetic GenesDespite the fact that pin and its glycosides biosynthesis pathway have remained obscure in E. ulmoides and Phomopsis sp. XP-8, our previous analysis indicates that detectable intermediates and corresponding enzyme activities exist, which are similar to those of the plant phenylpropanoid pathway in Phomopsis sp. XP-8[24]. To reveal the genetic basis of the pin and its glycosides biosynthesis pathway in Phomopsis sp. XP-8, functional annotation of genes using reference plant phenylpropanoid pathway genes was performed according to keyword searches for standard gene names and synonyms or abbreviations. Each search result was further confirmed with BLAST searches.
1.6 Construction of Phylogenetic Trees of Proteins Associated with Pinoresinol Formation and UDP GlucosyltransferaseDirigent proteins (DIR) or chloroperoxidase (CPO) and UDP Glucosyltransferase (UGT) are the two key enzymes involved in the pin and its glycoside derivatives biosynthetic pathways. However, comparison of CPO, DIR and UGT gene sequences in the genome examined with the sequences in the literature revealed no significant similarity. Therefore, protein searches were performed using the DIR, CPO, and UGT genes in the Phomopsis sp. XP-8 genome by BLASTp against GenBank databases from NCBI. Two phylogenetic trees were constructed using Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0.18. Protein sequence alignment (multiple alignments) was initially conducted by applying ClustalW (Gap opening penalty = 10, Gap extension penalty = 0.1) with default gap penalties. Phylogenetic trees were finally calculated based on the resulting alignments from ClustalW by using 1000 bootstrap steps.
1.7 Nucleotide Sequence Accession NumbersThe sequencing project was registered in the National Center for Biotechnology Information (NCBI) BioProject database with accession number PRJNA376863. Raw sequencing data, assembly, and annotations of Phomopsis sp. XP-8 genome have been deposited in Sequence Read Archive (SRA) database with accession number SRP100812.
2 Results 2.1 Genome Sequencing and General FeaturesThe assembled genome was generated into a total of 1617 contigs with a total length of 54384533 bp and N50 values of 59580 bp. The contigs were assembled into 330 scaffolds (≥1000 bp) with a total length of 55225560 bp and N50 values of 318484 bp. The genome features are shown in Table 1. The overall G+C% content is 53.50%, which is higher than the average G+C% content for fungi, which is 48.96%[45]. High G+C% content indicates greater thermal stability[46]. CEGMA analysis showed that 244 of 248 genes were fully annotated (98.39% completeness), and 246 of 248 genes met the criteria for partial annotation (99.19% completeness). BUSCO analysis indicated that 85% and 10% of 290 expected Ascomycota genes were identified as complete and fragmented; meanwhile, only 3% were missed in the assembly. Both assessment methods indicated that the Phomopsis sp. XP-8 genome assembly was fully complete. The genome contains 310 non-coding RNA (ncRNAs) (144 tRNA, 49 rRNA and 117 miRNA). Overall, 0.46 Mb of non-redundant and repetitive sequences were identified, comprising approximately 0.83% of the genome assembly.
| Genome | Gene number |
| Number of contigs | 1671 |
| Scaffolds (≥1000 bp) | 330 |
| Contigs length/bp | 54384533 |
| Scaffolds length/bp | 55225560 |
| Contigs N50/bp | 59580 |
| Scaffolds N50/bp | 318484 |
| Genome assembly size/Mb | 55.2 |
| G+C content (%) overall | 53.5 |
Gene prediction was performed using Augustus and yielded 17094 protein-coding genes. For all predicted genes, the functions of 16025 genes (93.75%) were successfully annotated in the different databases. Including 5876 genes > 300 nt and 9766 genes > 1000 nt. The functional annotation of each database is detailed below (Table 2). 15921 genes (93.14%) were aligned using the Nr database, 11893 genes (69.57%) had hits in the Swiss-Prot database, 2907 genes (17.01%) were mapped in the KEGG database, 6239 genes (36.50%) were classified in the GO database, 6788 genes (39.71%) were annotated against the COG database, the e-value thresholds were all set at 1e-5.
| Database | Number | Percentage/% |
| COG_Annotation | 6788 | 39.71 |
| GO_Annotation | 6319 | 36.97 |
| KEGG_Annotation | 3071 | 17.97 |
| KOG_Annotation | 8744 | 51.15 |
| Swissprot_Annotation | 11893 | 69.57 |
| Pfam_Annotation | 11556 | 67.60 |
| TrEMBL_Annotation | 15883 | 92.92 |
| nr_Annotation | 15921 | 93.14 |
| nt_Annotation | 9102 | 53.25 |
| Total | 16025 | 93.75 |
To further demonstrate the functional distribution of all genes, GO, COG and KEGG analyses were used for function prediction and classification. By GO analysis, the gene set was categorized into 45 functional groups (Figure 2). A total of 6319 genes were mapped to GO terms. The assignments were given to cellular components (17.14%), molecular functions (44.67%), and biological processes (38.19%). Among all of the GO terms, the vast majority were related to catalytic activity, metabolic processes, cell process, single-organism and binding. Within the cellular component category, cell part, cell and membrane occupied the majority of the genes. In the molecular function category, catalytic activity and binding proteins were dominant. Under the biological process category, metabolic process, single-organism process and cellular process represented the majorities, indicating that these genes were involved in some important metabolic activities in Phomopsis sp. XP-8.
|
| Figure 2 Gene ontology (GO) classification of Phomopsis sp. XP-8. The left-hand scale on the y-axis shows the percentage of genes in each category. The right-hand scale on the y-axis represents the number of genes in the same category. |
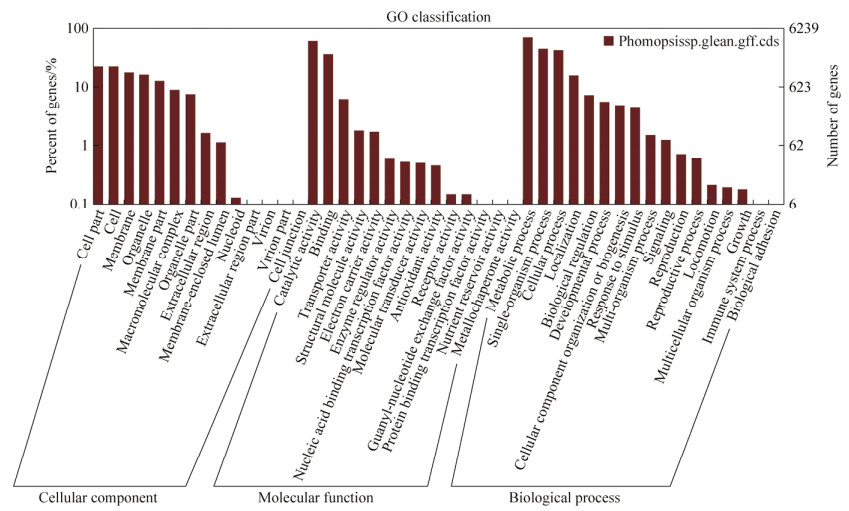
Further, all genes were searched against the COG database. Overall, 6788 sequences were assigned COG classifications. The COG-annotated putative proteins were classified into 25 functional categories (Figure 3). Among the 25 COG categories, the cluster for general function prediction only represented the largest one, followed by amino acid transport and metabolism, inorganic ion transport and metabolism. Notably, 905 (13.33%) genes were assigned to secondary metabolite biosynthesis, transport and catabolism, and 1074 (15.82%) were assigned to carbohydrate transport and metabolism. To identify the biological pathways in Phomopsis sp. XP-8, the annotated genes were mapped to the KEGG database (Figure 4). A total of 2907 genes were assigned to 116 KEGG pathways and classified into 5 main KEGG categories and 19 subcategories. Among them, the most represented were in the "metabolism pathways", followed by "biosynthesis of secondary metabolites". We focused considerably our attention on the biosynthesis of secondary metabolites (Table 3). 37 genes were identified in the phenylpropanoid pathway to verify that the genes of the pathway are related to lignan biosynthesis. Furthermore, 16 genes were mapped onto terpenoid backbone biosynthesis; 18 for stilbenoid, diaryl heptanoid and gingerol biosynthesis; 12 for plant- pathogen interaction; 16 for flavonoid biosynthesis; 4 for flavone and flavonol biosynthesis, and so on. These annotations would be anextremely valuable resource for further research on gene identification, mechanism for regulating gene expression, structures, functions, and metabolism pathways in Phomopsis sp. XP-8.
|
| Figure 3 Functional classification of the Clusters of Orthologous Groups (COGs). A total of 6788 genes were assigned to one or more of the 25 COG classification categories. |

|
| Figure 4 Functional classification of the Kyoto Encyclopedia of Genes and Genomes (KEGG). |
| Biosynthesis of secondary metabolites | Gene number | Pathway ID |
| Phenylpropanoid biosynthesis | 37 | ko00940 |
| Terpenoid backbone biosynthesis | 16 | ko00900 |
| Flavonoid biosynthesis | 16 | ko00941 |
| Flavone and flavonol biosynthesis | 4 | ko00944 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 18 | ko00945 |
| Isoquinoline alkaloid biosynthesis | 13 | ko00950 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 13 | ko00960 |
| Caffeine metabolism | 3 | ko00232 |
| Glucosinolate biosynthesis | 4 | ko00966 |
| Diterpenoid biosynthesis | 1 | ko00904 |
| Carotenoid biosynthesis | 35 | ko00906 |
| Limonene and pinene degradation | 24 | ko00903 |
2.2 Orthology and Phylogenetic Analysis
Using OrthoMCL, a total of 11818 gene families were identified in the Phomopsis sp. XP-8 genome. Among these gene families, 64 were unique. Compared with other five sequenced Ascomycota genomes, 12635 orthologous genes in Phomopsis sp. XP-8 are shared, whereas 1140 genes are unique for this fungus. 163 genes belong to unique gene families. Furthermore, the majority of 1140 species-unique genes are hypothetical proteins that do not contain recognizable Pfam domains, indicating unique gene functions. The annotated functions of species-unique gene proteins are mainly used in the biosynthesis of amino acids, aromatic amino acids metabolism, secondary metabolism and carbon metabolism processes. A Venn diagram shows that the largest cluster group has 5626 gene families each containing at least one gene from each of the six species. A phylogenomic tree that was generated based on 5042 single-copy orthologs revealed that Phomopsis sp. XP-8 and Diaporth ampelina (D. ampelina) are closely related to the Valsa mali (V. mali), which is consistent with a previous report[47] (Figure 5).
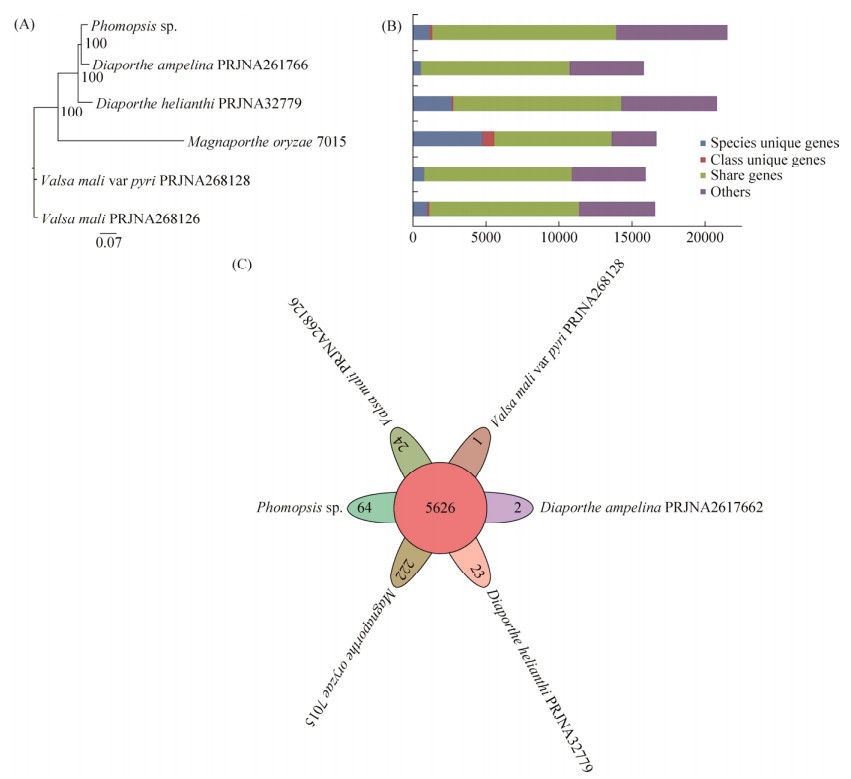
|
| Figure 5 Orthology gene clusters and phylogenetic analysis of Phomopsis sp. XP-8 and other five sequenced Ascomycota genomes. A: The maximum likelihood that the phylogenetic tree of the six Ascomycota species was created from 5042 single-copy orthologs using phyML with bootstrapping for 1000 replicates; B: The number of orthologous genes among the six Ascomycota species using OrthoMCL; C: A Venn diagram showing orthologous gene families among the six Ascomycota species. |
2.3 Considerable Biosynthetic Capabilities of Secondary Metabolites in Phomopsis sp. XP-8
Endophytic fungi may synthesize various bioactive secondary metabolites[48]. Phomopsis sp. XP-8 has been found to produce secondary metabolites[49], including pin and its glucosides[22].
This finding prompted us to determine the molecular basis of this production by genome sequencing. We detected 80 secondary metabolite gene clusters in Phomopsis sp. XP-8 by submitting the whole genome data to the antiSMASH automatic pipeline. The resulting enzymes included 30 Type Ⅰ polyketide synthases (T1PKS), 2 Type Ⅲ polyketide synthases (T3PKS), 16 non-ribosomal peptide synthases (NRPS), 7 terpene synthase (TS) group, 4 indolel groups, 2 lantipeptide group, 1 siderophore- indole group, 1 NRPS-T1PKS hybrid, 2 T1PKS- NRPS hybrids, 1 NRPS-T1PKS-indole hybrids, and 13 other groups. Among the 80 gene clusters, 53 were identified as PKS and NRPS (> 66% total). In addition, the core structure of secondary metabolites of the Phomopsis sp. XP-8 strain was also analyzed. 14 synthetic compound structures were predicted. These data, along with the obtained results in the numerous novel secondary metabolites, indicated a huge potential for the production of secondary metabolites of Phomopsis sp. XP-8.
2.4 Annotation of Pinoresinol and its Glycosides Biosynthetic GenesIn plants, lignan is normally produced by phenylpropanoid biosynthesis (ko00940). The genes involved in lignan synthesis have been isolated and characterized, such as PAL (DQ115905), C4H (GU014562), 4CL (GU937875), CCR (GQ872418), CAD (GU937874), C3H (JF826963), and CCoAOMT (DQ115905). The transcriptome database, by de novo techniques, provides a pool of candidate genes involved in the biosynthesis of phenylpropanoid in Isatis indigotica[50-52]. In the present study, we used the genes involved in the phenylpropanoid biosynthetic pathway in plants as a reference, for which 86 genes that encoded up to 11 enzymes were found be involved in this biosynthetic pathway for pin and its glycosides, including PAL, CYP73A, 4CL, shikimate O-hydroxycinnamoyltransferase (HCT), coumaroylquinate (coumaroylshikimate) 3′-monooxygenase (CYP98A), caffeoyl-CoA-O- methyltransferase (CCoAOMT), caffeic acid 3-O-methyltransferase (COMT), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), and CPO, DIR and UGT. All of these enzymes were successfully annotated and had more than one copy but PAL (Figure 6).

|
| Figure 6 Simplified pathways for the putative biosynthesis of pinoresinol and its glucoside derivatives in Phomopsis sp. XP-8. The Enzyme Commission (EC) number in red represents the enzyme of corresponding to the catalytic pathway. The names and numbers in brackets following each enzyme name in green indicate the abbreviation of the enzyme and the number of genes annotated in this pathway. |
2.5 Phylogenetic Analysis of Pinoresinol-Forming Associated genes
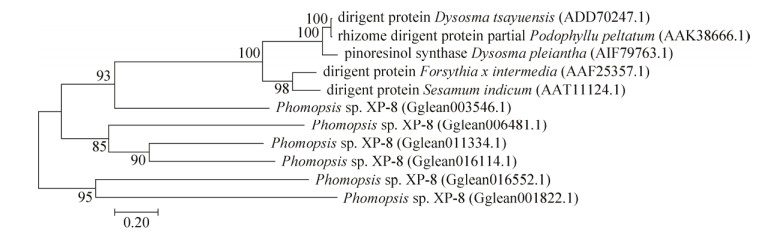
In the current study, the phylogenetic tree of the pin-forming associated proteins was generated from alignments of the full-length amino acid sequences from previously published data in GenBank and the proteins of the genome selected from Phomopsis sp. XP-8. As shown in Figure 7, Sesamum indicum (AAT11124.1), Forsythia x intermedia (AAF25357.1), Dysosma tsayuensis (ADD70247.1), Podophyllum peltatum (AAK38666.1), and Dysosma pleiantha (AIF79763.1) were the five pin-forming proteins in GenBank. These proteins had the same large clade, along with Gglean003546.1, which was annotated as the CPO gene in Phomopsis sp. XP-8. Gglean006481.1, Gglean011334.1, and Gglean016114.1 were three CPO genes with the same clade near the aforementioned clade. Gglean016552.1 and Gglean001822.1 comprised the two annotated dirigent protein genes and formed a separate large clade. Analyses indicated that in Phomopsis sp. XP-8, the pin was derived from the coupling of two coniferyl alcohol molecules that were more likely catalyzed by CPO than DIR. Although no reports exist that show that pin is catalyzed by the DIR or CPO in E. ulmoides and Phomopsis sp. XP-8, pin is formed by DIR in other plants.
|
| Figure 7 Phylogenetic tree of proteins associated with pinoresinol formation in Phomopsis sp. XP-8 and other known species proteins from NCBI. The phylogenetic tree was constructed using Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0.18. Protein sequence alignment (multiple alignments) was initially conducted by applying ClustalW (Gap opening penalty=10, Gap extension penalty=0.1) with default gap penalties. Phylogenetic trees were finally calculated based on the resulting alignments from ClustalW by using the neighbor-joining algorithm and 1000 bootstrap steps. |
2.6 Phylogenetic Analysis of Annotated UDP Glucosyltransferase
The phylogenetic analysis of UGTs clearly showed plants, fungi and Saccharomyces formed three separate and larger clades (Figure 8). Gglean016718.1 and several fungi (AQX36236.1, GAN01566.1, EPB92999.1, AAD29571.1), Saccharomyces (AAB67475.1 and AAD29570.1) were in the same large clade; Gglean004896.1, Gglean008180.1, Gglean002755.1, Gglean000988.1, Gglean014280.1, several plants, Eucommia ulmoides (AHX74090.1), Forsythia x intermedia (BAI65911.1 and BAI65912.1), and Sesamum alatum (BAF96581.1) were in another large clade, whereas Gglean007115.1, Gglean008006.1, Gglean013194.1, and Gglean011606.1 formed a separate clade. This analysis showed that Gglean016718.1 was closer to the previously mentioned fungi and Saccharomyces, which could be explored as a candidate gene for a study on glycosylation of pin glucoside derivatives.

|
| Figure 8 Phylogenetic tree of UGT protein in Phomopsis sp. XP-8 and other known species proteins from NCBI. The phylogenetic tree was constructed from a Clustal W multiple alignments using the neighbor-joining algorithm and the bootstrap value set to 1000 replicates by MEGA software version 7.0.18. |
The conserved domains database in NCBI was used to align the amino acid sequence of Gglean016718.1. The results showed that Gglean016718.1 was highly homologous to GT1-glycosyltransferase (GT1_Gtf_like, PSSM: cd03784), and the conserved domains were screened. There were 14 core amino acids (active site) involved, six of which were located at the TDP-binding site, one at the acceptor substrate-binding pocket. So, we infer the amino acid backbone of Gglean016718.1 is consistent with the characteristics of NDP- glycosyltransferase, and can be folded to form the corresponding active catalytic center and substrate binding sites.
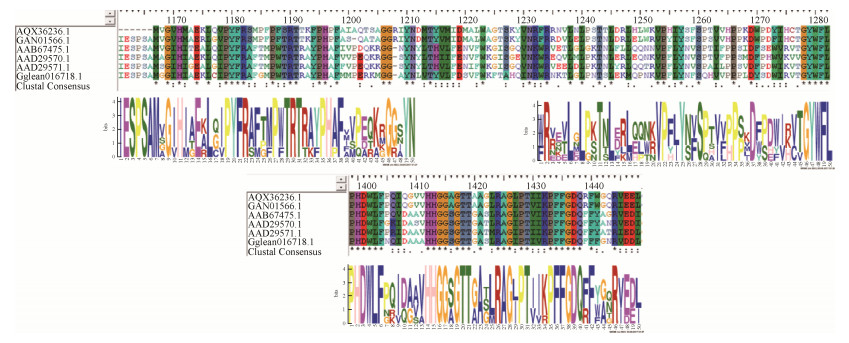
Gglean016718.1 had multiple sequences aligned with the corresponding sequences of glycosyltransferases in fungal and Saccharomyces proteins (Figure 9). Less than 50% identities were determined between Gglean016718.1 and Mucor hiemalis (AQX36236.1, 45%), Mucor ambiguous (GAN01566.1, 42%), Mucor circinelloides f. circinelloides 1006PhL Mucor ambiguous (EPB92999.1, 42%), Komagataella pastoris (AAD29570.1, 48%), Candida albicans (AAD29571.1, 46%), and Saccharomyces cerevisiae Ylr189cp (AAB67475.1, 46%). The amino acids at the positions of 1125-1174, 1190-1239 and 1351-1400 of Gglean016718.1 were three protein motifs found in the other six glycosyltransferases. These results suggested that Gglean016718.1 has the same biological function as that of fungal glycosyltransferases in the biosynthesis of phenols and sterol compounds.
|
| Figure 9 Multiple sequence alignments of Gglean016718.1 and protein MOTIF search. Amino acids were identified, conserved, and semi-conserved by comparison with other UGTs sequences from fungi in GenBank are indicated by black asterisks '*', one black dot '·', and two black dots ':', respectively. |
3 Discussion
Phomopsis was proposed to be an asexual state of Diaporthe[47]. As the phytopathogenic and endophytic species, Phomopsis have an extremely broad range of plant hosts and are widely distributed, including angiosperms, gymnosperms, pteridophytes, and bryophytes[53]. Phomopsis sp. are non-systematic fungi lacking host specificity and have been used as model endophytes in numerous studies[54]. As one class of the most widely distributed endophytic fungi, Phomopsis sp. has recently drawn increasing attention from the research community because of their ability to produce secondary metabolites with various structures and diverse biological activities[49, 55-58].
In plant, proteins associated with pin formation have been reported, which are classified as DIR. Related genes have been isolated and characterized in Sesamum indicum (AAT11124.1), Forsythia x intermedia (AAF25357.1), Dysosma tsayuensis (ADD70247.1), Podophyllum peltatum (AAK38666.1), and Dysosma pleiantha (AIF79763.1). However, Charles[19] in 1976 investigated a method of obtaining pin via the utilization of the precursor coniferyl alcohol and the catalyzation activity of CPO produced by Caldariomyces fumago. In plants, glycosylation represents the last step in the biosynthesis of numerous natural compounds. Glycosyltransferases family 1 (GT1), often referred to as a UGT, that is considerably important for chemical stability, water-soluble enhancement, inactivation, and detoxification of natural products. These enzymes generally catalyze the transfer of the glycosyl group from nucleoside diphosphate-activated sugars (UDP-sugars) to a diverse array of substrates, such as secondary metabolites or xenobiotics. The composition of lignans is enriched by glycoslation catalyzed by multiple UGTs. UGT71A18 serves as a (+)-pinoresinol glucosyltransferase in the Forsythia suspension culture that can produce PMG on HPLC analysis[59].
Pin, PDG and PMG have been detected as bioactive secondary metabolites of Phomopsis sp. XP-8[22]. DIR or CPO and UGTs are the two key enzymes involved in the pin and its glycoside biosynthesis. 2 DIR, 7 CPO and 14 UGT genes were annotated in the Phomopsis sp. XP-8 genome. To further screen these genes, annotation and identification of genes that code for carbohydrate-active enzymes (CAZymes) in the examined genome were performed using the CAZymes Analysis Toolkit (http://mothra.ornl.gov/cgi-bin/cat/cat.cgi). Combining genomic annotation and phylogenetic analysis, Gglean003546.1 and Gglean016718.1 will be further studied in the biosynthesis of pin and its glycosides as main candidate genes. This study provides genetic basis for the further gene cloning, gene expression, and functional verification, and may also identify more significant genes.
In addition, according to what has been found in plants, pin can be converted to many functional compounds, such as lariciresinol and secoisolariciresinol by pinoresinol/lariciresinol reductase and subsequently to matairesinol via secoisolariciresinol dehydrogenase (SIRD). In this study, 8 pinoresinol reductase genes, 5 pinoresinol-lariciresinol reductase genes, and 5 SIRD genes were found in Phomopsis sp. XP-8, indicating this strain might have the capability to synthesize other lignan compounds using pin as the precursor. Cytochalasins are other secondary metabolites produced by Phomopsis, and are derived from phenylalanine, tyrosine, leucine, tryptophan and alanine[60]. Schümann and Hertwerck[61] used RNA silencing to determine that the fungal PKS-NRPS hybrid synthase CheA plays an essential role in cytochalasan biosynthesis in Penicillium expansum. The genomic sequence analysis reveals that Phomopsis sp. XP-8 also has the potential to produce cytochalasan. Overall, the sequencing results indicate a huge potential for the production of a diverse array of industrially important natural products from Phomopsis sp. XP-8.
In conclusion, we report first genome sequencing, assembly, annotation and analysis of candidate genes involved in lignans and other secondary metabolites biosynthesis in the plant endophytic fungus Phomopsis sp. XP-8. This study provides basis for the further metabolic engineering of pin and its glucoside derivatives production, further studies into mining novel bioactive secondary metabolites of plant endophyte, and plant-endophyte interactions. More important, this research will provide important informations on the genes of Phomopsis sp. XP-8 and important reference to similar study of other endophytic fungi.
AcknowledgementsThis work was supported by the National Natural Science Foundation of China (Grant No. 31201408). The authors declare that they have no conflict of interests. We thank Biomarker Biotechnology Corporation (Beijing, China) for technical assistance with data analysis and Zhiyuan Yin, Ph.D. for suggestions about the data analysis.
| [1] | Zhao J, Mou Y, Shan T, Li Y, Zhou L, Wang M, Wang J. Antimicrobial metabolites from the endophytic fungus pichia guilliermondii isolated from Paris polyphylla var. yunnanensis. Molecules, 2010, 15(11): 7961-7970. DOI:10.3390/molecules15117961 |
| [2] | Sepporta MV, Mazza T, Morozzi G, Fabiani R. Pinoresinol inhibits proliferation and induces differentiation on human HL60 leukemia cells. Nutrition Cancer, 2013, 65(8): 1208-1218. DOI:10.1080/01635581.2013.828089 |
| [3] | Li Q, Zhang Y, Shi JL, Wang YL, Zhao HB, Shao DY, Huang QS, Yang H, Jin ML. Mechanism and anticancer activity of the metabolites of an endophytic fungi from eucommia ulmoides Oliv. Anti-Cancer Agents in Medicinal Chemistry, 2017, 17(7): 982-989. |
| [4] | Bomi HW, June Y, Qing H, Eun R, Dong G. Antifungal effect of (+)-pinoresinol isolated from Sambucus williamsii. Molecules, 2010, 15(5): 3507-3516. DOI:10.3390/molecules15053507 |
| [5] | During A, Debouche C, Raas T, Larondelle Y. Among plant lignans, pinoresinol has the strongest anti-inflammatory properties in human intestinal Caco-2 cells. Journal of Nutrition, 2012, 142(10): 1798-1805. DOI:10.3945/jn.112.162453 |
| [6] | Xie L, Akao T, Hamasaki K, Deyama T, Hattori M. Biotransformation of pinoresinol diglucoside to mammalian lignans by human intestinal micro flora, and isolation of Enterococcusfaecalis strain PDG-1 responsible for the transformation of (+)-pinoresinol to (+)-lariciresinol. Chemical and Pharmaceutical Bulletin, 2003, 51(5): 508-515. DOI:10.1248/cpb.51.508 |
| [7] | Wang CZ, Ma XQ, Yang DH, Guo ZR, Liu GR, Zhao GX, Tang J, Zhang YN, Ma M, Cai SQ, Ku BS, Liu SL. Production of enterodiol from defatted flaxseeds through biotransformation by human intestinal bacteria. BMC Microbiology, 2010, 10: 115. DOI:10.1186/1471-2180-10-115 |
| [8] | Wikul A, Damsud D, Kataoka K, Phuwapraisirisan P. (+)-Pinoresinol is a putative hypoglycemic agent in defatted sesame (Sesamum indicum) seeds through inhibiting α-glucosidase. Bioorganic & Medicinal Chemistry Letters, 2012, 22(16): 5215-5217. |
| [9] | Trantas E, Panopoulos N, Ververidis F. Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metabolic Engineering, 2009, 11(6): 355-366. DOI:10.1016/j.ymben.2009.07.004 |
| [10] | Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CCC, Oakley BR. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. Journal of The American Chemical Society, 2011, 133(11): 4010-4017. DOI:10.1021/ja1096682 |
| [11] | Wang B, Kang QJ, Lu YZ, Bai LQ, Wang CS. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(4): 1287-1292. DOI:10.1073/pnas.1115983109 |
| [12] | Chen L, Yue Q, Zhang XY, Xiang MC, Wang CS, Li SJ, Che YS, Ortiz-lópez FJ, Bills G, Liu XZ, An ZQ. Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis. BMC Genomics, 2013, 14: 339. DOI:10.1186/1471-2164-14-339 |
| [13] | Dixon RA, Achnine L, Kota P, Liu CJ, Reddy MSS, Wang LJ. The phenylpropanoid pathway and plant defence-a genomics perspective. Molecular Plant Pathology, 2002, 3(5): 371-390. DOI:10.1046/j.1364-3703.2002.00131.x |
| [14] | Jaini R, Wang P, Dudareva N, Chapple C, Morgan JA. Targeted metabolomics of the phenylpropanoid pathway in Arabidopsis thaliana using reversed phase liquid chromatography coupled with tandem mass spectrometry. Phytochemical Analysis, 2017, 28(4): 267-276. DOI:10.1002/pca.v28.4 |
| [15] | Verma VC, Kharmar RN, Strobel GA. Chemical and functional diversity of natural products from plant associated endophytic fungi. Natural Product Communications, 2009, 4(11): 1511-1532. |
| [16] | Ravindra PA, Suneel K, Sardul SS. Endophytic mycoflora as a source of biotherapeutic compounds for disease treatment. Journal of Applied Pharmaceutical Science, 2016, 6(10): 242-254. |
| [17] | He X, Wang J, Li M, Hao D, Yang Y, Zhang C, He R, Tao R. Eucommia ulmoides Oliv.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. Journal of Ethnopharmacology, 2014, 151(1): 78-92. DOI:10.1016/j.jep.2013.11.023 |
| [18] | Li C, Qiu G, Liu B, Chen J, Fu H. Neuroprotective effect of lignans extracted from Eucommia ulmoides Oliv. on glaucoma-related neurodegeneration. Neurological Sciences, 2016, 37(5): 755-762. DOI:10.1007/s10072-016-2491-3 |
| [19] | Charles JS, Ravikumt PR, Huang FC. Isolation and synthesis of pinoresinol diglucoside, a major antihypertensive principle of Tu-Chung (Eucommia ulmoides Oliv.). Journal of the American Chemical Society, 1976, 98(17): 5412-5413. DOI:10.1021/ja00433a070 |
| [20] | Maruyama J, Kobayashi M, Miyashita M, Kouno I, Irie H. A synthesis of (±)-pinoresinol and its related compound using potassium persulfate (K2S2O8) oxidation of benzoylacetates. Heterocycles, 1994, 37: 839-845. DOI:10.3987/COM-93-S72 |
| [21] | Esther R, Marco G, Vlada B. Three-steps in one-pot: whole-cell biocatalytic synthesis of enantiopure (+)- and (−)-pinoresinol via kinetic resolution. Microbial Cell Factories, 2016, 15: 78. DOI:10.1186/s12934-016-0472-0 |
| [22] | Zhang Y, Shi JL, Gao ZH, Yangwu R, Jiang H, Che JX, Liu YL. Production of pinoresinol diglucoside, pinoresinol monoglucoside, and pinoresinol by Phomopsis sp. XP-8 using mung bean and its major components. Applied Microbiology and Biotechnology, 2015, 99(11): 4629-4643. DOI:10.1007/s00253-015-6491-7 |
| [23] | Li C, Qiu G, Liu B, Chen J, Fu H. Neuroprotective effect of lignans extracted from Eucommia ulmoides Oliv. on glaucoma-related neurodegeneration. Neurological Sciences, 2016, 37(5): 755-762. DOI:10.1007/s10072-016-2491-3 |
| [24] | Zhang Y, Shi JL, Liu LP, Gao ZH, Che JX, Shao DY, Liu YL. Bioconversion of pinoresinol diglucoside and pinoresinol from substrates in the phenylpropanoid pathway by resting cells of Phomopsis sp. XP-8. PLoS ONE, 2015, 10(9): e0137066. DOI:10.1371/journal.pone.0137066 |
| [25] | Doroshenko VG, Livshits VA, Airich LG, Shmagina IS, Savrasova EA, Ovsienko MV, Mashko SV. Metabolic engineering of Escherichia coli for the production of phenylalanine and related compounds. Applied Biochemistry and Microbiology, 2015, 51(7): 733-750. DOI:10.1134/S0003683815070017 |
| [26] | Almakarem ASA, Heilman KL, Conger HL, Shtarkman YM, Rogers SO. Extraction of DNA from plant and fungus tissues in situ. BMC Research Notes, 2012, 5: 266. DOI:10.1186/1756-0500-5-266 |
| [27] | Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research, 2008, 18(5): 821-829. DOI:10.1101/gr.074492.107 |
| [28] | Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics, 2007, 23(9): 1061-1067. DOI:10.1093/bioinformatics/btm071 |
| [29] | Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 2015, 31(19): 3210-3212. DOI:10.1093/bioinformatics/btv351 |
| [30] | Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Research, 2007, 35: W265-W268. DOI:10.1093/nar/gkm286 |
| [31] | Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Research, 2010, 38(22): e199. DOI:10.1093/nar/gkq862 |
| [32] | Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics, 2005, 21(1): i351-i358. |
| [33] | Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics, 2005, 21(1): i152-i158. |
| [34] | Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, Sanmiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nature Reviewer Genetics, 2007, 8(12): 973-982. DOI:10.1038/nrg2165 |
| [35] | Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and Genome Research, 2005, 110(1-4): 462-467. DOI:10.1159/000084979 |
| [36] | Tarailo-Graovac M, Chen N. Using repeat masker to identify repetitive elements in genomic sequences. Current Protocols in Bioinformatics, 2009, 5(3): 4-14. |
| [37] | Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 1997, 25(5): 955-964. DOI:10.1093/nar/25.5.0955 |
| [38] | Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 1997, 25(17): 3389-3402. DOI:10.1093/nar/25.17.3389 |
| [39] | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Gavin SG. Gene ontology: tool for the unification of biology. Nature Genetics, 2000, 25(1): 25-29. DOI:10.1038/75556 |
| [40] | Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research, 2000, 28(1): 33-36. DOI:10.1093/nar/28.1.33 |
| [41] | Minoru K, Susumu G, Shuichi K. The KEGG resource for deciphering the genome. Nucleic Acids Research, 2004, 32: D277-D280. DOI:10.1093/nar/gkh063 |
| [42] | Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research, 2003, 13(9): 2178-2189. DOI:10.1101/gr.1224503 |
| [43] | Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics, 2003, 21(1): i152-i158. |
| [44] | Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systems Biology, 2010, 59(3): 307-321. DOI:10.1093/sysbio/syq010 |
| [45] | Li X, Du D. Variation, evolution, and correlation analysis of C+G content and genome or chromosome size in different kingdoms and phyla. PLoS ONE, 2014, 9(2): e88339. DOI:10.1371/journal.pone.0088339 |
| [46] | Zhou P, Zhang GQ, Chen SW, Jiang ZQ, Tang YB, Henrissat B. Genome sequence and transcriptome analyses of the thermophilic zygomycete fungus Rhizomucor miehei. BMC Genomics, 2014, 15: 294. DOI:10.1186/1471-2164-15-294 |
| [47] | Saccardo PA. Notae Mycologicae. Series Ⅴ Annales Mycologici, 1905, 3: 165-171. |
| [48] | Barbara S, Christine B, Siegfried D, Anne-Katrin R, Karsten K. Endophytic fungi: a source of novel biologically active secondary metabolites. Mycological Research, 2002, 106(9): 996-1004. DOI:10.1017/S0953756202006342 |
| [49] | Cai RL, Chen SH, Liu ZM, Tan CB, Huang XH, She ZG. A new α-pyrone from the mangrove endophytic fungus Phomopsis sp. HNY29-2B. Natural Product Research, 2017, 31(2): 124-130. DOI:10.1080/14786419.2016.1214833 |
| [50] | Lu BB, Du Z, Ding RX, Zhang L, Yu XJ, Liu CH, Chen WS. Cloning and characterization of a differentially expressed phenylalanine ammonia lyase gene (IilPAL) after genome duplication from tetraploid Isatis indigotica. Journal of Integrative Plant Biology, 2006, 48(12): 1439-1449. DOI:10.1111/jipb.2006.48.issue-12 |
| [51] | Hu YS, Di P, Chen JF, Xiao Y, Zhang L, Chen WS. Isolation and characterization of a gene encoding cinnamoyl-CoA reductase from Isatis indigotica. Molecular Biology Reports, 2011, 38(3): 2075-2083. DOI:10.1007/s11033-010-0333-6 |
| [52] | Chen JF, Dong X, Li Q, Zhou X, Gao SH, Chen RB, Sun LN, Zhang L, Chen WS. Biosynthesis of the active compounds of Isatis indigotica based on transcriptome sequencing and metabolites profiling. BMC Genomics, 2013, 14: 857. DOI:10.1186/1471-2164-14-857 |
| [53] | Gavin JA, Stodart B, Sakuanrungsirikul S, Anschaw E, Crump N. Anschaw E, Crump N. Genetic characterization of a novel Phomopsis sp., a putative biocontrol agent for Carthamus lanatus. Mycologia, 2010, 102(1): 54-61. DOI:10.3852/08-198 |
| [54] | Dai C, Chen Y, Tian L, Shi Y. Correlation between invasion by endophytic fungus Phomopsis sp. and enzyme production. African Journal of Agricultural Research, 2010, 5(11): 1324-1330. |
| [55] | Vatcharin R, Ubonta S, Souwalak P, Jariya S. Metabolites from the endophytic fungus Phomopsis sp. PSU-D15.. Phytochemistry, 2008, 69(3): 783-787. DOI:10.1016/j.phytochem.2007.09.006 |
| [56] | Yang J, Xu F, Huang C, Li J, She Z, Pei Z, Lin Y. Metabolites from the mangrove endophytic fungus Phomopsis sp. European Journal of Organic Chemistry, 2010, 19: 3692-3695. |
| [57] | Du G, Wang ZC, Hu WY, Yan KL, Wang XL, Yang HM, Yang HY, Gao YH, Liu Q, Hu QF. Three new 3-methyl-2- arylbenzofurans from the fermentation products of an endophytic fungus Phomopsis sp. and their anti-TMV activity. Phytochemistry Letters, 2017, 21: 287-290. DOI:10.1016/j.phytol.2016.04.003 |
| [58] | Wang M, Zhang W, Xu W, Shen Y, Du L. Optimization of genome shuffling for high-yield production of the antitumor deacetylmycoepoxydiene in an endophytic fungus of mangrove plants. Applied Microbiology and Biotechnology, 2016, 100(17): 7491-7498. DOI:10.1007/s00253-016-7457-0 |
| [59] | Eiichiro O, Hyun JK, Jun M, Kinuyo M, Atsushi O, Akio K, Toshiaki U, Honoo S. Molecular and functional characterization of novel furofuranclass lignan glucosyltransferases from Forsythia. Plant Biotechnology, 2010, 27(4): 317-324. DOI:10.5511/plantbiotechnology.27.317 |
| [60] | Zheng CJ, Shao CL, Wu LY, Chen M, Wang KL, Zhao DL, Sun XP, Chen GY, Wang CY. Bioactive phenylalanine derivatives and cytochalasins from the soft coral-derived fungus, Aspergillus elegans. Marine Drugs, 2013, 11(6): 2054-2068. DOI:10.3390/md11062054 |
| [61] | Schümann J, Hertwerck C. Molecular basis of cytochalasan biosynthesis in fungi: gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. Journal of the American Chemical Society, 2007, 129(31): 9564-9565. DOI:10.1021/ja072884t |
 2018, Vol. 58
2018, Vol. 58