中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 刘波, 唐洁, 陈廷廷, 曾林, 曾朝懿, 张庆. 2018
- Bo Liu, Jie Tang, Tingting Chen, Zeng Lin, Chaoyi Zeng, Qing Zhang. 2018
- 一株路德维希肠杆菌的筛选及其对3-苯氧基苯甲酸的降解特性分析
- Screening and characterization of a 3-phenoxybenzoic acid degrading Enterobacter ludwigii
- 微生物学报, 58(5): 830-841
- Acta Microbiologica Sinica, 58(5): 830-841
-
文章历史
- 收稿日期:2017-05-16
- 修回日期:2017-07-05
- 网络出版日期:2017-08-04
2. 西华大学食品与生物工程学院, 古法发酵生物技术研究所, 四川 成都 610039
2. Institute of Ancient Brewing, School of Food and Biotechnology, Xihua University, Chengdu 610039, Sichuan Province, China
拟除虫菊酯类农药(pyrethroid pesticides)是一类人工合成的模拟天然除虫菊素的仿生型杀虫剂[1],在防治果蔬、茶叶、谷物类害虫以及蚊蝇、蟑螂和牲畜寄生虫等上有显著作用。然而拟除虫菊酯类农药具有较强的疏水性,在自然条件下仅存在少量降解,其余大部分在施用后直接吸附并富集于水和土壤等环境中[2],易被动植物吸收;而人体内的拟除虫菊酯类农药残留约90%来自于受污染的水源、植物或动物性食物,这在尿液中拟除虫菊酯类农药代谢产物[3-苯氧基苯甲酸(3-phenoxybenzoic acid,3-PBA)、二氯菊酸等]的监测结果中得到确认[3-4]。相较于拟除虫菊酯类农药对环境和动植物造成的直接污染和毒害作用,其中间代谢产物所造成的二次污染更值得重视[1, 3]。其中3-PBA是绝大多数拟除虫菊酯类农药(如溴氰菊酯、氯氰菊酯、氯菊酯和氰戊菊酯等)的必经降解中间产物之一[5-6]。最新研究发现,3-PBA是潜在的环境雌激素类物质[7],具有生殖毒性[8];且其疏水性相对母体农药较弱[9],自然条件下难降解,易迁移、蓄积[10],半衰期可达180 d[11],潜在危害性更大。因此,如何消除3-PBA残留是解决环境中拟除虫菊酯类农药污染的关键。
目前,受拟除虫菊酯类农药污染环境的修复研究主要集中于物理催化法、化学氧化法和生物降解法等[10]。与物理和化学法相比,生物降解法以其条件温和、降解效率高、无二次污染等特点[2],已成为目前解决环境中农药残留问题的热点方法[1],因此筛选出高效持久的3-PBA降解菌株是环境中拟除虫菊酯类农药残留去除的关键。现国内外报道的3-PBA降解菌主要来源于废水排污淤泥、农药污染土壤和酱油曲料中,如从活性淤泥中分离出的鞘氨醇单胞菌(Sphingomonas sp.)[12]、假单胞菌(Pseudomonas sp.)[13]、苍白杆菌(Ochrobactrum lupini)[14]、江苏鞘氨醇杆菌(Sphingobium jiangsuense)[15]和嗜麦芽寡养单胞菌(Stenotrophomonas sp.)[16],在农药污染土壤中筛选得到的芽孢杆菌(Bacillus sp.)[17],以及从酱油曲料中分离的米曲霉(Aspergillus oryzae)[18]等,这些筛选得到的3-PBA降解菌均具有较好的3-PBA利用能力以及生物修复受拟除虫菊酯类农药污染环境的潜力。但其中大多数降解菌株对3-PBA的耐受浓度有限,难以应用于3-PBA高污染环境的修复治理;而相对于拟除虫菊酯类农药或其他污染物的降解菌株[19-20],现有关于3-PBA降解菌株的报道甚少,且未见从植物源土壤中分离出3-PBA高效降解菌株的报道,所以迫切需要从植物源土壤中筛选具有植物亲附能力的3-PBA高效降解菌株,以期为环境中3-PBA或拟除虫菊酯类农药残留的生物降解提供菌株资源。
本文报道从受拟除虫菊酯类农药污染的大豆根系土壤中分离出1株路德维希肠杆菌(Enterobacter ludwigii),对该菌的生长降解动力学、3-PBA降解特性进行分析,并优化降解条件,为其进一步应用于受3-PBA或拟除虫菊酯类农药污染环境的生物修复提供理论基础。
1 材料和方法 1.1 材料与仪器 1.1.1 样品来源:本实验所采用的样品采自川北地区某种植基地的大豆根系、根际土壤样品,采样深度为5-20 cm。
1.1.2 试剂与培养基:3-苯氧基苯甲酸(3-phenoxybenzoic acid,3-PBA),梯希爱(上海)化学化工发展有限公司;乙腈(色谱纯),美国Adamas-beta公司;Bacteria DNA Kit提取试剂盒、多重PCR扩增试剂盒,天根生化科技有限公司;基础盐培养基(mineral salt medium,MSM;g/L):(NH4)2SO4 1.5、KH2PO4 0.5、K2HPO4 1.5、MgSO4 0.2、NaCl 0.5,pH 7.5,121 ℃灭菌20 min,3-PBA浓度根据试验所需配制;结晶紫中性红胆盐琼脂(violet red bile agar,VRBA)培养基41.5 g,蒸馏水1000 mL,煮沸2 min,冷却倒平板备用;液体培养基加入2.0% (W/V)的琼脂制成固体培养基。
1.1.3 仪器:Waters e2695型高效液相色谱仪、Waters 2998型光电二极管阵列检测器,美国Waters公司;Biometra PCR仪,德国Analytik Jena AG公司;JSM7500F型扫描电镜,日本电子株式会社;UNICO 7200型紫外分光光度计,美国UNICO公司;Heraeus Multifuge X1R高速冷冻离心机,美国thermo Scientific公司。
1.2 3-PBA的检测及其标准曲线方程建立3-PBA检测采用高效液相色谱法(high performance liquid chromatography,HPLC)进行。样品处理和HPLC检测方法参照文献[12]进行,其中流动相为乙腈/超纯水(70︰30,V/V)。标准曲线方程:配制3-PBA标准液(100 mg/L),依次进样1、2、5、10、20 μL,拟合3-PBA质量-峰面积的标准曲线为y=89.3658x+0.7803,R2=1.0000。
1.3 3-PBA降解菌株的筛选与鉴定3-PBA降解菌的富集驯化和筛选纯化参照文献[21]进行,驯化过程的3-PBA终浓度为1600 mg/L。纯化后的菌株转接到含3-PBA (100 mg/L)的MSM培养基中,接入同体积无菌水做空白对照,30 ℃振荡(180 r/min)培养48 h后取样测定3-PBA残留量,并按式(1)计算3-PBA降解率,筛选高效降解菌株进行后续研究。

|
公式(1) |
C:样品组中3-PBA含量;C0:对照组中3-PBA含量。
降解菌株形态、生理生化特征、总DNA提取、16S rRNA扩增及序列测定和系统发育树的构建参照文献[21]进行。其中,16S rRNA扩增体系包括引物27f (AGAGTTTGATCCTGGCTCAG) 0.5 μL,1492r (GGTTACCTTGTTACGACTT) 0.5 μL,DNA模板1 μL,ddH2O 10.5 μL,混合酶(含Taq酶、dNTPs、MgSO4等) 12.5 μL。
1.4 降解菌的生长降解动力学将菌株种子液(OD600=1.0)[17]按5.0% (V/V)的接种量分别接种到含3-PBA (100 mg/L)的MSM培养基中,30 ℃振荡(180 r/min)培养72 h,定时取样,测定其吸光度(optical density,OD)值和3-PBA残留量。
1.4.1 菌体生长动力学模型:在某一特定条件下,微生物生长的最大细胞浓度与底物浓度有关[22],因此采用Logistic方程描述菌体生长动力学模型。在此方程中比生长速率与尚未利用的负载能力相关联,即生长期细胞的增长速率可以用(2)式表达。

|
公式(2) |
当t=t0,X=X0时,将(2)式积分得到(3)式。

|
公式(3) |
X:菌体浓度(OD600);t:培养时间(h);μm:最大比生长速率(h-1);X0:初始菌体浓度(OD600);Xm:最大菌体浓度(OD600)。
1.4.2 3-PBA降解动力学模型:酶促反应动力学方程被广泛应用于酶促反应分析,主要研究酶催化的反应速率及影响反应速率的各种因素[23]。其中,一级动力学方程在污染物的生物降解研究中被广泛应用,如(4)式所示。

|
公式(4) |
当S远小于Ks时,v=vmaxS/Ks,即v=dS/dt=vmax/Ks×S=-k1S,积分得:ln S=-k1t+ln S0,变形得(5)式。
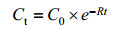
|
公式(5) |
C0:3-PBA初始浓度(mg/L);Ct:t时刻3-PBA浓度(mg/L);k:降解速率常数(h-1);t:降解时间(h)。
按(6)式计算底物降解半衰期t1/2(h)。

|
公式(6) |
将种子液(OD600=1.0)以5%的接种量接种至含3-PBA的MSM培养基中,在不同的培养温度(30、34、37、40 ℃)、初始pH (5.5、6.5、7.5、8.5)及3-PBA浓度(25、50、100、200 mg/L)条件下180 r/min振荡培养,接入等量生理盐水作空白对照。每隔8 h取样,测定3-PBA残留量,并以一级降解动力学方程进行拟合,分析降解菌的降解特性及最适降解条件。
1.6 Box-Behnken响应曲面设计优化条件根据1.5实验的结果,选取响应面设计的中心点。利用Design-Expert 8.0软件,采用Box-Behnken中心组合设计3因素3水平的响应曲面实验;对实验数据进行统计和分析,确定菌株降解3-PBA的最佳降解条件,并在此条件下培养48 h后,测定3-PBA残留量,计算降解率。
2 结果和分析 2.1 3-PBA降解菌的筛选鉴定 2.1.1 降解菌株的筛选及纯化:经富集驯化、分离纯化共获得23株可耐受3-PBA (1600 mg/L)的菌株,其中8株具备有效降解3-PBA的能力,培养48 h对3-PBA (100 mg/L)的降解能力如图 1所示。菌株BPBA031降解能力最好,达到55.04%,经方差分析可知菌株BPBA031降解率与其他菌株降解率差异性显著(P<0.01)。表明菌株BPBA031是1株3-PBA高效降解菌,故选用该菌株做后续研究。

|
| 图 1 不同菌株对3-PBA的降解率 Figure 1 Degredation rate of 3-PBA by different strains. Different letters indicate significant difference, P < 0.01. |
2.1.2 降解菌株的形态及生理生化特征:
菌株BPBA031纯化后分别划线接种于MSM、VRBA培养基中,其在MSM培养基上呈圆形,乳白色,中央微凸,湿润粘稠有光泽,直径2-4 mm;在VRBA培养基上呈圆形或椭圆形,紫红色边缘红色沉淀,湿润粘稠有明亮光泽,直径3-8 mm,有胆盐水解圈(1-3 cm);菌体(图 2)呈短圆杆状,单个排列,两端较平整,大小为(0.6-1.0) μm×(0.8-1.8) μm。根据菌株的形态特征可初步预测降解菌BPBA031属于肠杆菌科细菌。

|
| 图 2 菌株BPBA031扫描电镜形态 Figure 2 Scanning electron microscopic morphology of strain BPBA031. |
菌株BPBA031生理生化实验结果见表 1。3-PBA降解菌BPBA031为革兰氏阴性兼性厌氧菌,发酵葡萄糖、乳糖,V-P反应、赖氨酸脱羧酶实验均呈阳性,吲哚和硫化氢实验呈阴性,表明菌株BPBA031极可能属于肠杆菌属细菌[24-25]。
| Test | Culture time/h | |
| 24 | 72 | |
| Gram stain | - | - |
| Aerobic | ± | ± |
| Sucrose fermentation | ++ | +++ |
| Glucose fermentation | +++ | +++ |
| Maltose fermentation | +++ | +++ |
| Lactose fermentation | +++ | +++ |
| Indole production | - | - |
| Methyl red test | +++ | +++ |
| Catalase | +++ | +++ |
| Urease activity | - | - |
| Citrate | + | +++ |
| NO3- reductase | +++ | +++ |
| Gelatin hydrolysis | +++ | +++ |
| Lysine decarboxylase | ++ | +++ |
| Voges-Proskauer | +++ | +++ |
| Denitrification | - | - |
| H2S production | - | - |
| β-galactosidase | +++ | +++ |
| Myo-inositol | +++ | +++ |
| α-methyl-D-glucoside | +++ | +++ |
| Galactitol | - | - |
| Ribitol | - | - |
| -: 0%-10%; ±: 10%-20%; +: 20%-80%; ++: 80%-90%; +++: 90%-100%. | ||
2.1.3 分离菌株16S rRNA序列分析:
将菌株BPBA031的16S rRNA扩增产物回收测序获得1395 bp的序列,并将此序列提交至GenBank (Accession number:KY474547)。利用BLAST检索分析将该序列进行同源性比对,发现菌株BPBA031与Enterobacter ludwigii R6-346-1 (Accession number:JQ659806)、Enterobacter ludwigii RCB553 (Accession number:KT260765)的同源性达99%,可以推测菌株BPBA031为路德维希肠杆菌(Enterobacter ludwigii)。为进一步确定菌株BPBA031的种属地位,利用MEGA 6.0软件及其Neighbor-Joining法构建系统发育树(图 3),菌株BPBA031与Enterobacter ludwigii R6-346-1、Enterobacter ludwigii RCB553处于同一分支,这也证实了菌株BPBA031为路德维希肠杆菌(Enterobacter ludwigii)。

|
| 图 3 菌株BPBA031基于16S rRNA的菌株系统发育分析 Figure 3 Phylogenetic tree of strain BPBA031 based on 16S rRNA sequence. The number in parentheses is accession number of GenBank; node number represents the confidence level of relatives; the length of branch represents the evolutionary distance and the coefficient is 0.01. |
综合形态学、生理生化特征[24, 26]、16S rRNA基因序列和系统发育分析,可以确定菌株BPBA031为路德维希肠杆菌(Enterobacter ludwigii)。
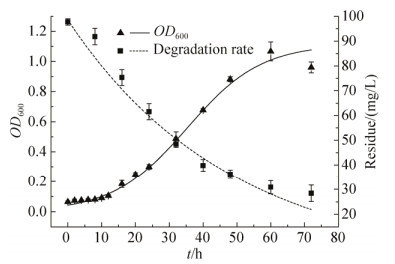
2.2 菌株BPBA031的生长降解动力学如图 4所示,利用Origin 8.0软件对菌株生物量实测值与菌体生长动力学模型进行非线性拟合,得到μm=0.09149 h-1,X0=0.04262及Xm=1.1145,代入(6)式得到菌株生长动力学方程:X=0.04262e0.09149t/[1-0.03824(1-0.04262e0.0915t)];利用一级反应动力学模型对降解过程中的3-PBA残留量进行非线性拟合,得到菌株对3-PBA的一级降解动力学方程:Ct=98.5583×e-0.02085t,k=0.02085;t1/2=33.24 h,表明路德维希肠杆菌BPBA031在以3-PBA为唯一碳源的MSM培养基中生长态势良好。在生长延滞期(0-10 h),适量3-PBA被利用于菌体生长和初始繁殖,生长对数期(10-48 h)菌体繁殖迅速,3-PBA降解速率达到最高,在生长稳定期对3-PBA利用明显减弱。生长降解动力学模型的拟合度分别为0.9824、0.9865,表明2种模型能较好反映路德维希肠杆菌BPBA031的实际生长规律以及对3-PBA的降解过程。
|
| 图 4 路德维希肠杆菌BPBA031生长和降解曲线 Figure 4 Growth and degradation curve of strain Enterobacter ludwigii BPBA031. |
2.3 菌株BPBA031的降解特性
采用一级降解动力学模型对不同条件下的3-PBA残留量进行拟合,得到不同培养温度、3-PBA浓度及初始pH条件下的3-PBA降解曲线(图 5)。

|
| 图 5 不同培养条件下3-PBA残留量变化 Figure 5 Degradation curves of 3-PBA at different culture conditions by strain BPBA031. A: Culture temperature; B: 3-PBA concentration; C: Initial pH. |
由不同培养温度条件下[3-PBA (100 mg/L)、初始pH 7.5]的降解曲线可知(图 5-A),菌株在30-40 ℃条件下对3-PBA的降解作用均比较显著;其中34 ℃条件下降解率为67.89%,t1/2=29.38 h;37 ℃条件下降解率为62.21%,t1/2=34.16 h。不同3-PBA浓度条件下(37 ℃,初始pH 7.5)的降解结果显示(图 5-B),菌株对不同浓度的3-PBA降解作用明显;其中浓度为50 mg/L时,3-PBA的降解率达75.77% (t1/2=24.95 h);低浓度(25 mg/L)及高浓度(100、200 mg/L)条件下的降解率稍低(t1/2分别为27.24 h、35.99 h及40.35 h)。如图 5-C所示,当初始pH介于5.5-8.5之间[37 ℃,3-PBA (100 mg/L)],菌株在初始pH 7.5时降解效果最好(降解率为66.44%,t1/2=30.75 h),初始pH 8.5条件下次之(降解率为52.87%,t1/2=39.93 h);而当初始pH为5.5、6.5时,3-PBA有降解但利用率较低。
2.4 菌株BPBA031降解3-PBA的条件优化采用Box-Behnken响应曲面试验设计,以培养温度(X1)、3-PBA浓度(X2)和初始pH(X3)为自变量,3-PBA降解率(Y)为响应值,根据2.3中实验结果确定响应面分析中心点(表 2),其他条件分别为MSM培养基、菌龄24 h、接种量5% (V/V,OD600=1.0)、转速180 r/min,得到路德维希肠杆菌BPBA031降解条件的优化试验设计及响应值(表 3)。
| Variables | Coded | Range and levels | ||
| -1 | 0 | 1 | ||
| Incubation temperature/℃ | X1 | 34.0 | 37.0 | 40.0 |
| 3-PBA concentration /(mg/L) |
X2 | 30.0 | 50.0 | 70.0 |
| Initial pH | X3 | 7.0 | 7.5 | 8.0 |
| Run | X1/℃ | X2/(mg/L) | X3 | Degradation rate/% |
| 1 | 0 | 0 | 0 | 78.25 |
| 2 | 0 | 0 | 0 | 76.88 |
| 3 | 0 | 1 | -1 | 53.44 |
| 4 | 0 | 0 | 0 | 78.96 |
| 5 | 0 | -1 | 1 | 65.56 |
| 6 | 1 | 0 | -1 | 41.26 |
| 7 | -1 | -1 | 0 | 81.11 |
| 8 | -1 | 0 | 1 | 66.64 |
| 9 | 0 | -1 | -1 | 55.89 |
| 10 | 0 | 0 | 0 | 79.38 |
| 11 | 1 | 0 | 1 | 38.56 |
| 12 | 0 | 1 | 1 | 62.09 |
| 13 | -1 | 0 | -1 | 53.15 |
| 14 | 0 | 0 | 0 | 77.88 |
| 15 | 1 | 1 | 0 | 58.55 |
| 16 | -1 | 1 | 0 | 75.25 |
| 17 | 1 | -1 | 0 | 60.56 |
采用Design-Expert 8.0软件对表中的实验数据进行多项式回归分析建立二次响应回归模型,拟合得到3-PBA降解率(Y)响应曲面模型:

|
方差分析的显著性检验结果(表 4)表明,模型F值(185.50)大于F0.01 (9.5),说明模型在F0.01水平上显著;X1X3交互项P值(<0.01)小于0.05,表明在该模型下培养温度与初始pH 2因素交互项显著;失拟项P值(0.16)大于0.05,即失拟项不显著,且决定系数R2=0.9958,表明拟合度较好。
| Source | Sum of squares | Df | Mean square | F value | P-value |
| Model | 2928.92 | 9 | 325.44 | 185.50 | <0.01 |
| X1 | 547.70 | 1 | 547.70 | 312.19 | <0.01 |
| X2 | 7.91 | 1 | 7.91 | 4.51 | 0.07 |
| X3 | 75.65 | 1 | 75.65 | 43.12 | <0.01 |
| X1X2 | 3.71 | 1 | 3.71 | 2.11 | 0.19 |
| X1X3 | 65.53 | 1 | 65.53 | 37.35 | <0.01 |
| X2X3 | 0.26 | 1 | 0.26 | 0.15 | 0.71 |
| X12 | 369.87 | 1 | 369.87 | 210.83 | <0.01 |
| X22 | <0.01 | 1 | <0.01 | <0.01 | 0.96 |
| X32 | 1519.20 | 1 | 1519.20 | 865.96 | <0.01 |
| Residual | 12.28 | 7 | 1.75 | ||
| Lack of fit | 8.49 | 3 | 2.83 | 2.98 | 0.16 |
| Pure error | 3.79 | 4 | 0.95 | ||
| Cor total | 2941.20 | 16 |
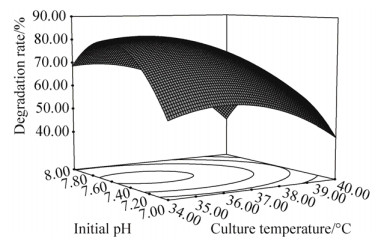
如图 6所示,当3-PBA浓度为30 mg/L时,接种量和初始pH交互作用对3-PBA降解率影响明显。可以看出,随着培养温度和初始pH的增加,3-PBA的降解率随之增大;增加到一定值后,其降解率逐步下降。
|
| 图 6 培养温度与初始pH交互作用的响应面图和等高线图 Figure 6 Response surface and contours plots of the interaction between culture temperature and initial pH. |
分别对模型的各自变量求一阶偏导,得其极值点:X1=-0.6,X2=-1.0,X3=0.17。即在35.19 ℃、3-PBA (30.00 mg/L)及初始pH 7.58条件下,路德维希肠杆菌BPBA031对3-PBA的降解率为83.48%;重复验证实验结果显示,优化条件下的3-PBA平均降解率为83.75%,与预测值基本一致,表明此模型能够准确预测不同培养条件下路德维希肠杆菌BPBA031对3-PBA的降解率。
3 讨论3-PBA是典型的拟除虫菊酯类农药降解残留物,具有毒性大、易蓄积以及难降解等特点;而生物降解具有条件温和、经济高效且不易形成二次污染等优势,因此可采用生物修复方法治理受3-PBA或拟除虫菊酯类农药污染的环境[27]。本课题组首次从大豆根系土壤中分离得到具有3-PBA高效降解能力的菌株BPBA031,综合形态学、生理生化特征、16S rRNA基因序列和系统发育分析的结果,将其鉴定为路德维希肠杆菌(Enterobacter ludwigii)。路德维希肠杆菌(Enterobacter ludwigii)是由Hoffmann等[28]首次从临床样品中分离得到,这种细菌曾一直被认为是人类病原体[29],但近年研究表明其不仅来自临床样品,在其他领域也被陆续发现。如Kwon等[30]在韩国发酵蔬菜中分离出高产粘性胞外多糖的Enterobacter ludwigii GK-23;从植物根际土壤或根结节中也筛选得到Enterobacter ludwigii,其大多具有植物生长促进作用[26]、产胞外多糖[31]、生物控制能力以及烃降解能力[32];Enterobacter ludwigii也存在于石油管道中的腐蚀微生物群落中[33];而Singh等[34]则从迁徙山羊的瘤胃内发现产单宁鞣酸酶的Enterobacter ludwigii GRT-1;因此Enterobacter ludwigii在食品工业、生物医疗、生物化工、植物促生定殖及环境治理等领域都有非常突出的应用价值[26, 31-35]。
同时,菌株BPBA031对3-PBA耐受浓度达1600 mg/L,远高于Halden等[13]和Chen等[14]从活性淤泥中分离出的Pseudomonas pseudoalcaligenes POB310和Ochrobactrum lupini DG-S-01;且该菌接种48 h对3-PBA (30 mg/L)降解率达83.75%,高于Zhu等[18]从酱油曲料中筛选出的Aspergillus oryzae M4;表明菌株BPBA031是1株高效3-PBA降解菌,同时具有生物修复受高浓度3-PBA或拟除虫菊酯类农药污染环境的潜力。此外,基于Logistic生长动力学和一级降解动力学模型拟合的生长降解动力学曲线表明,菌株在延滞期(0-10 h)对3-PBA的利用较为明显,进入对数期(10-48 h) 3-PBA则被迅速降解,但进入稳定期(48 h后)时间较早,可能是由于碳源的缺乏或是底物的反馈抑制作用所造成,这与Chen等[17]的研究结果比较一致;且3-PBA的半衰期(t1/2=33.24 h)远低于Stenotrophomonas sp. ZS-S-01[16];表明该菌在以3-PBA为唯一碳源的培养条件下,可迅速吸收利用3-PBA。而菌株BPBA031在不同培养条件下对3-PBA的降解效率差异表明:(1)菌株在34-37 ℃对3-PBA降解作用明显(图 5-A),推测菌株分泌的3-PBA降解酶在此温度范围内的培养体系中具有最佳催化活性;(2)高浓度3-PBA在一定程度上可抑制菌株对3-PBA的降解(图 5-B),但结果表明强抑制作用可能出现在较高的3-PBA浓度范围,鉴于污染环境中3-PBA或拟除虫菊酯类农药残留浓度远低于实验条件[1, 27],推测菌株在自然环境条件下对3-PBA应具有较高降解效率;(3)在较酸或较碱条件下,微生物表面及其生长环境的电荷可能发生改变[36],进而影响微生物对3-PBA的吸收和降解利用;而且3-PBA降解后会生成酸性产物[17, 37],导致pH值降低,因此推测中性和稍偏碱性的环境更有利于菌株对3-PBA的降解,这与本次实验结果(图 5-C)相一致。
本研究表明从受拟除虫菊酯类农药污染的大豆根系土壤中分离的路德维希肠杆菌(Enterobacter ludwigii) BPBA031菌株可高效降解3-PBA,具有潜在的植物促生特性以及芳烃降解能力,可作为生物修复受3-PBA或拟除虫菊酯类农药污染环境的优良微生物资源。
| [1] | Cycoń M, Piotrowska-Seget Z. Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: a review. Frontiers in Microbiology, 2016, 7: 1463. |
| [2] | Zhang C, Jia L, Wang SH, Qu J, Li K, Xu LL, Shi YH, Yan YC. Biodegradation of beta-cypermethrin by two Serratia spp. with different cell surface hydrophobicity. Bioresource Technology, 2010, 101(10): 3423-3429. DOI:10.1016/j.biortech.2009.12.083 |
| [3] | Chuang JC, Van Emon JM, Trejo RM, Durnford J. Biological monitoring of 3-phenoxybenzoic acid in urine by an enzyme-linked immunosorbent assay. Talanta, 2011, 83(5): 1317-1323. DOI:10.1016/j.talanta.2010.07.077 |
| [4] | Watkins DJ, Fortenberry GZ, Sánchez BN, Barr DB, Panuwet P, Schnaas L, Osorio-Valencia E, Solano-González M, Ettinger AS, Hernández-Ávila M, Hu H, Téllez-Rojo MM, Meeker JD. Urinary 3-phenoxybenzoic acid (3-PBA) levels among pregnant women in Mexico City: distribution and relationships with child neurodevelopment. Environmental Research, 2016, 147: 307-313. DOI:10.1016/j.envres.2016.02.025 |
| [5] | Sun H, Chen W, Xu XL, Ding Z, Chen XD, Wang XR. Pyrethroid and their metabolite, 3-phenoxybenzoic acid showed similar (anti) estrogenic activity in human and rat estrogen receptor α-mediated reporter gene assays. Environmental Toxicology and Pharmacology, 2014, 37(1): 371-377. DOI:10.1016/j.etap.2013.11.031 |
| [6] | McCoy MR, Yang Z, Fu X, Ahn KC, Gee SJ, Bom DC, Zhong P, Chang D, Hammock BD. Monitoring of total type Ⅱ pyrethroid pesticides in citrus oils and water by converting to a common product 3-phenoxybenzoic acid. Journal of Agricultural and Food Chemistry, 2012, 60(20): 5065-5070. DOI:10.1021/jf2051653 |
| [7] | Liu Y, Wu AH, Hu J, Lin MM, Wen MT, Zhang X, Xu CX, Hu XD, Zhong JF, Jiao LX, Xie YJ, Zhang CZ, Yu XY, Liang Y, Liu XJ. Detection of 3-phenoxybenzoic acid in river water with a colloidal gold-based lateral flow immunoassay. Analytical Biochemistry, 2015, 483(1): 7-11. |
| [8] | Yuan C, Wang C, Gao SQ, Kong TT, Chen L, Li XF, Song L, Wang YB. Effects of permethrin, cypermethrin and 3-phenoxybenzoic acid on rat sperm motility in vitro evaluated with computer-assisted sperm analysis. Toxicology in vitro, 2010, 24(2): 382-386. DOI:10.1016/j.tiv.2009.11.001 |
| [9] | Sun H, Xu XL, Xu LC, Song L, Hong X, Chen JF, Cui LB, Wang XR. Antiandrogenic activity of pyrethroid pesticides and their metabolite in reporter gene assay. Chemosphere, 2007, 66(3): 474-479. DOI:10.1016/j.chemosphere.2006.05.059 |
| [10] | Topp E, Akhtar MH. Mineralization of 3-phenoxybenzoate by a two-membered bacterial co-culture. Canadian Journal of Microbiology, 1990, 36(7): 495-499. DOI:10.1139/m90-086 |
| [11] | Vidal JLM, Plaza-Bolaños P, Romero-González R, Frenich AG. Determination of pesticide transformation products: a review of extraction and detection methods. Journal of Chromatography A, 2009, 1216(40): 6767-6788. DOI:10.1016/j.chroma.2009.08.013 |
| [12] | Tang J, Yao K, Liu SL, Jia DY, Chi YL, Zeng CY, Wu S. Biodegradation of 3-phenoxybenzoic acid by a novel Sphingomonas SP. SC-1. Fresenius Environmental Bulletin, 2013, 22(5): 1564-1572. |
| [13] | Halden RU, Tepp SM, Halden BG, Dwyer DF. Degradation of 3-phenoxybenzoic acid in soil by Pseudomonas pseudoalcaligenes POB310(pPOB) and two modified Pseudomonas strains. Applied and Environmental Microbiology, 1999, 65(8): 3354-3359. |
| [14] | Chen SH, Hu MY, Liu JJ, Zhong GH, Yang L, Rizwan-ul-Haq M, Han HT. Biodegradation of beta-cypermethrin and 3-phenoxybenzoic acid by a novel Ochrobactrum lupini DG-S-01. Journal of Hazardous Materials, 2011, 187(1/3): 433-440. |
| [15] | Zhang J, Lang ZF, Zheng JW, Hang BJ, Duan XQ, He J, Li SP. Sphingobium jiangsuense sp. nov., a 3-phenoxybenzoic acid-degrading bacterium isolated from a wastewater treatment system. International. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(Pt 4): 800-805. DOI:10.1099/ijs.0.029827-0 |
| [16] | Chen SH, Yang L, Hu MY, Liu JJ. Biodegradation of fenvalerate and 3-phenoxybenzoic acid by a novel Stenotrophomonas sp. strain ZS-S-01 and its use in bioremediation of contaminated soils. Applied Microbiology and Biotechnology, 2011, 90(2): 755-767. DOI:10.1007/s00253-010-3035-z |
| [17] | Chen SH, Hu W, Xiao Y, DengYY, Jia JW, Hu MY. Degradation of 3-phenoxybenzoic acid by a Bacillus sp. PLoS One, 2012, 7(11): e50456. DOI:10.1371/journal.pone.0050456 |
| [18] | Zhu YT, Li JL, Yao K, Zhao N, Zhou K, Hu XJ, Zou LK, Han XF, Liu AP, Liu SL. Degradation of 3-phenoxybenzoic acid by a filamentous fungus Aspergillus oryzae M-4 strain with self-protection transformation. Applied Microbiology and Biotechnology, 2016, 100(22): 9773-9786. DOI:10.1007/s00253-016-7847-3 |
| [19] | Wang Y, Chen H, Liu YX, Ren RP, Lv YK. An adsorption-release-biodegradation system for simultaneous biodegradation of phenol and ammonium in phenol-rich wastewater. Bioresource Technology, 2016, 211: 711-719. DOI:10.1016/j.biortech.2016.03.149 |
| [20] | Pinyakong O, Habe H, Supaka N, Pinpanichkarn P, Juntongjin K, Yoshida T, Furihata K, Nojiri H, Yamane H, Omori T. Identification of novel metabolites in the degradation of phenanthrene by Sphingomonas sp. strain P2. FEMS Microbiology Letters, 2000, 191(1): 115-121. DOI:10.1111/fml.2000.191.issue-1 |
| [21] |
Liu B, Tang J, Chen TT, Shi Y, Zeng L, Zeng CY. Isolation and identification of two pyrethroid pesticides degradation strains and research of degradative capabilities. Science and Technology of Food Industry, 2017, 38(4): 214-219, 224.
(in Chinese) 刘波, 唐洁, 陈廷廷, 史颖, 曾林, 曾朝懿. 两株拟除虫菊酯类农药降解菌的分离鉴定及其降解能力的研究. 食品工业科技, 2017, 38(4): 214-219, 224. |
| [22] |
Zhang YX, Meng XJ, Chai TY. Characterization of phenol-degrading Rhodococcus sp. strain P1 from coking wastewater. Acta Microbiologica Sinica, 2013, 53(10): 1117-1124.
(in Chinese) 张玉秀, 蒙小俊, 柴团耀. 苯酚降解菌红球菌(Rhodococcus sp.)P1的鉴定及其在焦化废水中的应用. 微生物学报, 2013, 53(10): 1117-1124. |
| [23] |
Ren L, Shi YH, Jia Y, Yao XS, Nahurira R, Mi CX, Yan YC. Biodegradation characteristics and kinetics of p-nitrophenol by strain Arthrobacter sp. CN2. Environmental Science, 2015, 36(5): 1757-1762.
(in Chinese) 任磊, 史延华, 贾阳, 姚雪松, NahuriraR, 弥春霞, 闫艳春. 菌株Arthrobacter sp.CN2降解对硝基苯酚的特性与动力学. 环境科学, 2015, 36(5): 1757-1762. |
| [24] | Hoffmann H, Stindl S, Ludwig W, Stumpf A, Mehlen A, Heesemann J, Monget D, Schleifer KH, Roggenkamp A. Reassignment of Enterobacter dissolvens to Enterobacter cloacae as E. cloacae subspecies dissolvens comb. nov. and emended description of Enterobacter asburiae and Enterobacter kobei. Systematic and Applied Microbiology, 2005, 28(3): 196-205. DOI:10.1016/j.syapm.2004.12.010 |
| [25] |
Lu LL, Xiao M, Xu XD. Enterobacter agglomerans B1 producing β-galactosidase with transglycosylation activity: screening, identification, fermentation conditions, and galacto-oligosaccharides synthesis. Acta Microbiologica Sinica, 2008, 48(1): 38-44.
(in Chinese) 卢丽丽, 肖敏, 徐晓东. 转糖基β-半乳糖苷酶产生菌Enterobacter agglomerans B1:筛选鉴定、发酵产酶及其合成低聚半乳糖的研究. 微生物学报, 2008, 48(1): 38-44. |
| [26] | Shoebitz M, Ribaudo CM, Pardo MA, Cantore ML, Ciampi L, Curá JA. Plant growth promoting properties of a strain of Enterobacter ludwigii isolated from Lolium perenne rhizosphere. Soil Biology and Biochemistry, 2009, 41(9): 1768-1774. DOI:10.1016/j.soilbio.2007.12.031 |
| [27] | Haritash AK, Kaushik CP. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. Journal of Hazardous Materials, 2009, 169(1/3): 1-15. |
| [28] | Hoffmann H, Stindl S, Stumpf A, Mehlen A, Monget D, Heesemann J, Schleifer KH, Roggenkamp A. Description of Enterobacter ludwigii sp. nov., a novel Enterobacter species of clinical relevance. Systematic and Applied Microbiology, 2005, 28(3): 206-212. DOI:10.1016/j.syapm.2004.12.009 |
| [29] | Paauw A, Caspers MPM, Schuren FHJ, Hall MALV, Delétoile A, Montijn RC, Verhoef J, Fluit AC. Genomic diversity within the Enterobacter cloacae complex. PLoS One, 2008, 3(8): e3018. DOI:10.1371/journal.pone.0003018 |
| [30] | Kwon TY, Shim SM, Heo MY, An DH, Shin KS, Lee JH. Isolation and characterization of exopolysaccharide-producing bacteria from Korean fermented vegetables. Korean Journal of Microbiology and Biotechnology, 2007, 35(3): 191-195. |
| [31] | Pau-Roblot C, Lequart-Pillon M, Apanga L, Pilard S, Courtois J, Pawlicki-Jullian N. Structural features and bioremediation activity of an exopolysaccharide produced by a strain of Enterobacter ludwigii isolated in the Chernobyl exclusion zone. Carbohydrate Polymers, 2013, 93(1): 154-162. DOI:10.1016/j.carbpol.2012.09.025 |
| [32] | Yousaf S, Afzal M, Reichenauer TG, Brady CL, Sessitsch A. Hydrocarbon degradation, plant colonization and gene expression of alkane degradation genes by endophytic Enterobacter ludwigii strains. Environmental Pollution, 2011, 159(10): 2675-2683. DOI:10.1016/j.envpol.2011.05.031 |
| [33] | Neria-González I, Wang ET, Ramírez F, Romero JM, Hernández-Rodríguez C. Characterization of bacterial community associated to biofilms of corroded oil pipelines from the southeast of Mexico. Anaerobe, 2006, 12(3): 122-133. DOI:10.1016/j.anaerobe.2006.02.001 |
| [34] | Singh B, Bhat TK, Sharma OP, Kanwar SS, Rahi P, Gulati A. Isolation of tannase-producing Enterobacter ludwigii GRT-1 from the rumen of migratory goats. Small Ruminant Research, 2012, 102(2/3): 172-176. |
| [35] | Yang M, Cai J, Wang CG, Du X, Lin JG. Characterization of endo-β-mannanase from Enterobacter ludwigii MY271 and application in pulp industry. Bioprocess and Biosystems Engineering, 2017, 40(1): 35-43. DOI:10.1007/s00449-016-1672-z |
| [36] |
Li HX, Lin H, You SH, Xu XY, Xia SQ. Effect of pH on reductive degradation of para-chloronitrobenzene by autohydrogenotrophic microorganisms. Acta Scientiae Circumstantiae, 2015, 35(7): 2083-2089.
(in Chinese) 李海翔, 林华, 游少鸿, 徐晓茵, 夏四清. pH对氢基质自养微生物还原降解对氯硝基苯的影响. 环境科学学报, 2015, 35(7): 2083-2089. |
| [37] | Maloney SE, Maule A, Smith AR. Microbial transformation of the pyrethroid insecticides: permethrin, deltamethrin, fastac, fenvalerate, and fluvalinate. Applied and Environmental Microbiology, 1988, 54(11): 2874-2876. |
 2018, Vol. 58
2018, Vol. 58