中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 张德福, 赵禹宗, 刘东, 冯娇, 张明, 曲道峰, 周冬生, 励建荣, 殷喆, 王丽. 2018
- Defu Zhang, Yuzong Zhao, Dong Liu, Jiao Feng, Ming Zhang, Daofeng Qu, Dongsheng Zhou, Jianrong Li, Zhe Yin, Li Wang. 2018
- 宋内志贺氏菌1173质粒基因组及耐药基因转移机制
- Bacterial plasmid genome analysis and the mechanism of resistance gene transfer in the clinical Shigella sonnei isolate 1173
- 微生物学报, 58(3): 467-475
- Acta Microbiologica Sinica, 58(3): 467-475
-
文章历史
- 收稿日期:2017-05-08
- 修回日期:2017-05-25
- 网络出版日期:2017-07-21
2. 军事医学科学院微生物流行病研究所, 病原微生物生物安全国家重点实验室, 北京 100071;
3. 河南大学第一附属医院检验科, 河南 开封 475000;
4. 浙江工商大学食品与生物工程学院, 浙江 杭州 310018
2. State Key Laboratory of Pathogen and Biosecurity, Institute of Microbiology and Epidemiology, Academy of Military Medical Sciences, Beijing 100071, China;
3. Department of Clinical Laboratory, the First Hospital Affiliated to Henan University, Kaifeng 475000, Henan Province, China;
4. School of Food Science and Biotechnology, Zhejiang Gongshang University, Hangzhou 310018, Zhejiang Province, China
Li Wang, Tel/Fax: +86-371-22736871, E-mail:wangli851217@163.com
志贺氏菌属(Shigellae)通称痢疾杆菌,是广泛存在于自然环境中的革兰氏阴性病原菌,其引起的细菌性腹泻是常见的一种胃肠道疾病[1-2]。据报道,全球每年约有1.6亿人因感染志贺氏菌患病,约占腹泻病例的15%以上[3]。根据血清型的不同,志贺氏菌可以分为福氏志贺氏菌(S. flexneri)、宋内志贺氏菌(S. sonnei)、鲍氏志贺氏菌(S. boydii)和痢疾志贺氏菌(S. dysenteriae)四种[1]。志贺氏菌属菌群分布复杂,因国家、地区和年代的不同而有差异,且世界各地流行的志贺氏菌每隔20-30年发生一次变迁[2]。发达国家以宋内志贺氏菌为主,发展中国家以福氏志贺氏菌为主[4];在我国,以福氏志贺氏菌和宋内志贺氏菌多见,且由福氏志贺氏菌向宋内志贺氏菌转移的趋势[5]。由于抗菌药物在临床及畜牧养殖业中的广泛应用导致的选择性压力使大量多药耐药宋内志贺氏菌出现[6-7],之前有效的青霉素类、四环素类和磺胺类药物不再有效,临床用药的形势变得严峻起来,严重威胁公众健康[8-9]。
产β-内酰胺酶是革兰氏阴性菌耐药的重要方式[10]。自上个世界八十年代早期大量使用有广谱抗菌作用的头孢菌素以来,超广谱β-内酰胺酶(Extended-spectrum beta-lactamase,ESBL)开始出现。它可以水解包括第三代头孢菌素和单环β-内酰胺类在内的广谱β-内酰胺类抗菌药物,并使之失去活性[10-11]。CTX-M已成为目前世界上最常见的超广谱β-内酰胺酶类型,并且已发现存在于肠杆菌科多个种属细菌可接合转移的质粒上,从而能够在同种属和不同种属的细菌间水平转移[11]。目前,CTX-M型β-内酰胺酶根据氨基酸序列的差异可以分为至少6个类群:CTX-M-1、CTX-M-2、CTX-M-8、CTX-M-9、CTX-M-25和KLUC。其中,CTX-M-45、CTX-M-64、CTX-M-123和CTX-M-132四个亚型是类群间的杂合体[12]。
本研究中,我们对临床分离到的1株产ESBL宋内志贺氏菌1173的耐药表型进行了鉴定,确定耐药质粒能够通过接合转移水平转移给其他细菌,并提取质粒进行了高通量基因组序列测定及生物信息学分析,对携带耐药基因的移动元件和耐药基因座位进行了精细注释,以探明其耐药基因转移的机制。
1 材料和方法 1.1 材料 1.1.1 菌株: 菌株1173分离自2011年就诊于302医院的一位65岁女性腹泻患者的大便样本。经沙门氏菌-志贺氏菌(Salmonella-Shigella,SS)培养基分离,挑单菌落进行玻片凝集实验鉴定,然后经质谱仪(Bruker公司)和16S rRNA基因扩增,最终鉴定为宋内志贺氏菌。 1.1.2 主要试剂: Taq DNA聚合酶、DNA分子量标准为生工生物工程(上海)股份有限公司产品,Pfu酶和dNTPs购自Fermentas公司,Qiagen Large Construct Kit购自QIAGEN公司,引物由北京博迈德基因技术有限公司合成,头孢他啶(Ceftazidime,CAZ) (30 μg)、头孢他啶/克拉维酸(Clavulanic acid,CA) (30 μg/10 μg)、头孢噻肟(Cefotaxime,CTX) (30 μg)和头孢噻肟/克拉维酸(30 μg/10 μg)药敏纸片购自OXID公司,MH、BHI培养基购自BD Bioscience公司,SS培养基购自北京陆桥技术股份有限公司。 1.2 ESBL确证实验ESBL确证实验参照美国临床和实验室标准协会(CLSI)标准[13],分别将头孢他啶和头孢他啶/克拉维酸纸片、头孢噻肟和头孢噻肟/克拉维酸纸片贴于涂布菌液的MH平板表面。37 ℃孵育18-20 h后观察、测量抑菌圈大小。含酶抑制剂的抑菌圈直径与单药抑菌圈相比≥5 mm时,即判定该菌株产ESBL。
1.3 耐药基因扩增及CTX-M全序列测定根据Chen等[14]的报道,合成主要的ESBL基因和碳青霉烯酶基因扩增引物,稀释到浓度为10 μmol/L备用。根据试剂盒的说明,提取1173的全基因组DNA,稀释为10 μmol/L的工作液,-20 ℃保存备用或直接作为模板进行靶基因扩增,扩增产物经1%浓度的琼脂糖电泳后置于紫外灯下观察拍照,PCR产物送博迈德公司进行序列测定。PCR的反应体系为:DNA 10 ng,10× Buffer 5 μL,dNTPs 0.5 μL,Taq酶2 U,Pfu酶0.5 U,0.1 μmol/L上下游引物各0.5 μL,ddH2O 3 μL。PCR的反应程序为:95 ℃ 5 min;94 ℃ 40 s,52 ℃ 30-50 s,72 ℃ 50 s,30个循环;72 ℃ 5 min。
以ISEcp1-F1 (5′-GGAGTGTATGAAAAATGT CTGG-3′)/orf477-R1 (5′-CGTGCTGTCGCTGGATA CC-3′)为引物对进行blaCTX-M-orf477区全序扩增及序列测定,与GenBank中的参考序列进行对比,确定blaCTX-M的基因亚型。
1.4 接合转移实验根据蒋晓圆等[15]的方法,以blaCTX-M阳性的1173为供体菌,以利福平耐药大肠杆菌EC600 (LacZ-,NalR,RifR)为受体菌进行接合转移实验。将受体菌和供体菌分别接种于BHI肉汤中过夜培养,分别取3 mL供体菌和受体菌混合,4 ℃下3000×g离心5 min,加入80 μL BHI肉汤吹打悬浮,点加于贴在BHI琼脂培养基的滤膜上(1 cm2大小,0.45 μm孔径),37 ℃倒置培养12-18 h。取滤膜用适量BHI肉汤培养基(200 μg/mL氨苄西林,2.5 mg/mL利福平)将菌体洗下,涂布于含有200 μg/mL氨苄西林和2.5 mg/mL利福平的BHI琼脂培养基上,37 ℃倒置培养24-28 h后的单菌落即为接合子(1173-CTXM-EC600)。以ISEcp1-F1/ orf477-R1为引物对,PCR鉴定接合子是否存在blaCTX-M基因,利用质谱及16S rRNA基因扩增测定接合子是否为受体菌,并对接合子进行双纸片协同扩散法的ESBL确证实验。
1.5 MIC测定采用VITEK® 2仪器进行MIC的检测(法国梅里埃公司),并按照CLSI[13]文件标准进行判定。
1.6 质粒基因组序列测定及生物信息学分析将宋内志贺氏菌1173中携带blaCTXM的质粒命名为p1173-CTXM,用Qiagen Large Construct Kit提取接合子1173-CTXM-EC600的质粒DNA,用Illumina HiSeq 2500进行高通量全基因组鸟枪法测序,并对GenBank中的数据进行BLASTn比对分析,查找近缘质粒,并以近缘质粒的序列为参考用Velvet软件进行序列拼接,然后采用PCR和Sanger双脱氧链终止法测序技术对缺失部分进行补充,获得质粒序列全长。对全序列利用GeneMarkS和RAST对全序列进行基因预测,随后通过对NR库、UniProtKB/Swiss-Prot等数据库进行比对分析,对预测的编码区基因进行注释。移动元件利用ISfinder、INTEGRALL及Tn number registry网站进行预测分析,并进行精细注释。使用Inkscape v0.48软件进行质粒图谱的绘制。
1.7 序列登录号将p1173-CTXM全序列及注释结果提交至GenBank,登录号为KY174331。
2 结果和分析 2.1 ESBL确证实验双纸片协同扩散法结果表明(图 1),1173对头孢噻肟/克拉维酸的抑菌圈为35 mm,对头孢噻肟的抑菌圈为10 mm,相差25 mm≥5 mm;对头孢他啶/克拉维酸的抑菌圈为30 mm,对头孢他啶的抑菌圈为21 mm,相差9 mm≥5 mm。因此,1173确证产ESBL。同理,1173-CTXM-EC600亦产ESBL。
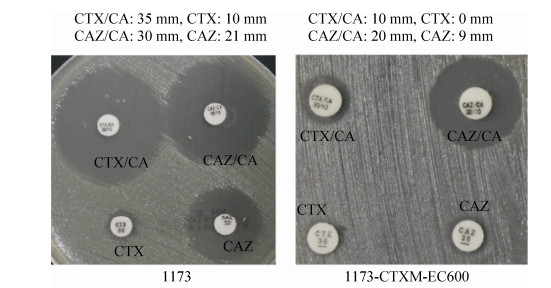
|
| 图 1 双纸片协同扩散实验 Figure 1 Double-disk synergy test. Each experiment was conducted with three parallel replicates: three plates inoculated with three independent bacterial cultures of strains 1173 and its transconjugant strains p1173-CTXM-EC600, respectively. Only a representative figure is shown here. CA: clavulanic acid; CAZ: ceftazidime; CTX: cefotaxime. |
2.2 耐药基因扩增及CTX-M全序列测定
利用PCR方法对1173及接合子1173-CTXM-EC600进行ESBL基因及碳青霉烯耐药基因的鉴定结果表明(图 2),1173及其接合子1173-CTXM-EC600都能扩增出blaCTX-M-1G基因,且PCR扩增产物片段大小与理论值一致。利用ISEcp1-F1/ orf477-R1引物进一步扩增及序列分析表明,该基因亚型为blaCTX-M-64。此外,1173还含有blaTEM基因,而其接合子无blaTEM基因。这表明,1173只有blaCTX-M-64能够通过接合转移进入受体菌。
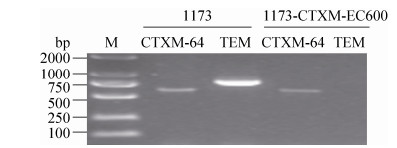
|
| 图 2 ESBL及碳青霉烯酶基因PCR扩增结果 Figure 2 PCR detection of bla genes. Strains 1173 and p1173-CTXM-EC600 were screened for the major ESBL and carbapenemase genes with PCR. Of all the bla genes tested, blaCTX-M-64, blaTEM were present in 1173, but only blaCTX-M-64 was detected in p1173-CTXM-EC600. |
2.3 MIC测定结果
采用VITEK® 2仪器对菌株1173及1173-CTXM-EC600的MIC检测结果(表 1)表明,二者均对氨苄西林、氨苄西林/舒巴坦、氨曲南、头孢唑啉、头孢曲松、头孢吡肟耐药,均对亚胺培南、厄他培南、环丙沙星、左氧氟沙星、呋喃妥因和替加环素敏感;而对头孢西丁、阿米卡星、庆大霉素、妥布霉素和复方新诺明,1173均表现为耐药,1173-CTXM-EC600表现为敏感。
| Category | Antibiotics | MIC (mg/L)/antimicrobial susceptibility | ||
| 1173 | 1173-CTXM-EC600 | EC600 | ||
| Penicillins | Ampicillin | ≥32R | ≥32R | 16I |
| Ampicillin/sulbactam | ≥32R | ≥32R | 4S | |
| Monobactams | Aztreonam | 32R | ≥64R | ≤1S |
| Cephalosporins | Cefazolin | ≥64R | ≥64R | ≤4S |
| Ceftriaxone | ≥64R | ≥64R | ≤1S | |
| Cefoxitin | ≤4R | 8S | 8S | |
| Cefepime | ≥64R | ≥64R | ≤1S | |
| Carbapenems | Imipenem | ≤1S | ≤1S | ≤1S |
| Ertapenem | ≤0.5S | ≤0.5S | ≤0.5S | |
| Fluoroquinolones | Ciprofloxacin | 0.5S | ≤0.25S | ≤0.25S |
| Levofloxacin | 2S | ≤0.5S | 0.5S | |
| Furanes | Levofloxacin | ≤16S | ≤16S | ≤16S |
| Aminoglycosides | Amikacin | ≤2R | ≤2S | ≤2S |
| Gentamicin | ≥16R | ≤1S | ≤1S | |
| Tobramycin | 4R | ≤1S | ≤1S | |
| Sulfanilamides | Trimethoprim/sulfamethoxazole | ≥320R | ≤20S | ≤20S |
| Tetraciclinas | Tigecycline | 1S | ≤0.5S | ≤0.5S |
| S: susceptible; I: intermediate; R: resistant. | ||||
2.4 质粒基因组序列测定及生物信息学分析
对质粒p1173-CTXM的高通量测序结果表明,该质粒全长为63530 bp,平均G+C%含量为42.3%,预测有90个开放阅读框(ORF) (图 3),平均基因长度为706 bp。将p1173-CTXM的序列与GenBank的nr/nt核酸数据库进行BLASTn比对分析表明,p1173-CTXM与IncI2型质粒p1081-CTXM (GenBank登录号KJ460501)[16]和pHN1122-1 (GenBank登录号JN797501)[17]为近缘质粒,其覆盖率(Coverage)和一致性(Identity)分别为97%和99%。对p1173-CTXM质粒序列进行生物信息学分析表明,p1173-CTXM的结构可以分为骨架区(Backbone)和外源插入区(Accessory modules)两部分。骨架区含有保守的IncI2质粒骨架区基因,又可进一步分为质粒复制区(Plasmid replication)、质粒稳定区(Plasmid maintenance)和质粒接合转移区(Conjugal transfer)。质粒复制区编码与质粒复制相关的质粒复制起始蛋白RepR/RepA,质粒稳定区编码与质粒稳定相关的Mok/Hok蛋白等,质粒接合转移区编码与质粒接合转移相关的NikABC和菌毛蛋白Pil/Tra等。
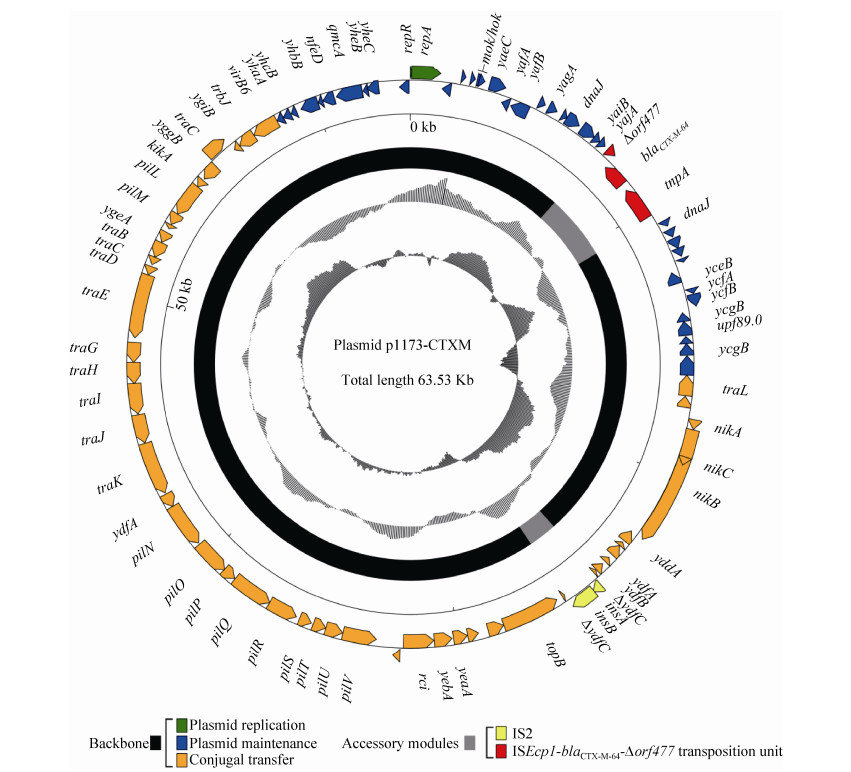
|
| 图 3 质粒p1173-CTXM全序列示意图 Figure 3 Schematic map of the sequence of plasmid p1173-CTXM. Genes are denoted by arrows and colored according to gene function classification. The innermost circle presents GC skew ([G-C]/[G+C]) with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content. The backbone and accessory module regions are also shown. |
p1173-CTXM的外源插入区由插入序列ISEcp1和其携带的耐药基因blaCTX-M-64组成的ISEcp1-blaCTX-M-64-Δorf477转座单元和插入序列IS2组成。ISEcp1-blaCTX-M-64-Δorf477转座单元长度为3080 bp,两端为同源重组形成的5 bp正向重复序列(Direct repeat,DR)。ISEcp1由两端的14 bp反向重复序列[Inverted repeat left (IRL) (5′-GGAT CTAAGATGCA-3′)和Inverted repeat right (IRR-1) (5′-ACGTGGAATTTAGG-3′)]和1263 bp的转座酶(Transposase,tnpA)基因组成。
p1173-CTXM还携带有2个近缘质粒所没有的IS2;此外,与p1081-CTXM和pHN1122-1相比,p1173-CTXM分别有1.1 kb和约500 bp的片段发生了颠转(图 4)。
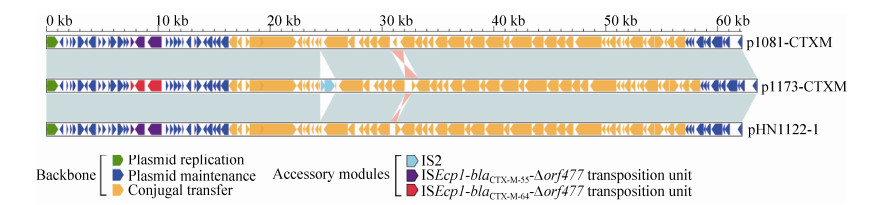
|
| 图 4 p1173-CTXM与近缘质粒序列线性比较 Figure 4 Linear comparison of sequenced plasmid p1173-CTXM. Genes are denoted by arrows colored according to gene function classification. Shaded regions denote regions of homology (> 95% nucleotide identity). |
3 讨论
目前,已报道的CTX-M基因已有180余种亚型(http://www.ncbi.nlm.nih.gov/pathogens/beta-lactamase-data-resources),根据氨基酸序列的异质性可以分为6个群,其中Group 1和Group 9是世界范围内最主要的流行群。blaCTX-M-64隶属于Group 1,是blaCTX-M-55和blaCTX-M-14的杂合体[18]。已报道的宋内志贺氏菌携带的CTX-M至少有10种亚型,如CTX-M-2、CTX-M-3、CTX-M-14、CTX-M-15、CTX-M-55和CTX-M-64等,而在中国,常见的类型为CTX-M-14、CTX-M-15和CTX-M-55[16]。blaCTX-M通常由插入序列ISEcp1和ISCR1或噬菌体相关序列携带,形成ISEcp1-blaCTX-M-IS903D或ISEcp1-blaCTX-M-orf477转座单元,在相同或不同种属的肠杆菌科细菌间进行水平转移、传播[11]。
ISEcp1属于IS1380家族,与普通的插入序列不同,ISEcp1为单末端插入序列,它仅靠单个插入序列即可介导其下游邻近的耐药基因进行转移,从而大大提高了耐药基因转移的概率[19-20]。在p1173-CTXM的ISEcp1-blaCTX-M-64-Δorf477转座单元中,ISEcp1通过识别其自身的IR和转座单元下游另外一个与IRR-1序列高度相似的IRR-2 (5′-ACGTAGGTCCCAGG-3′)介导ISEcp1-blaCTX-M-64-Δorf477转座单元整体在细菌间的传播[16, 19]。ISEcp1的IRR-1与blaCTX-M-64的起始密码子ATG间有45 bp间隔区(Spacer)。常见的间隔区还有42 bp和127 bp等形式,其长短可影响blaCTX-M-64的表达[21]。对GenBank中的blaCTX-M-64序列分析表明,CTX-M-64质粒的宿主菌范围比较窄。目前已报道的携带CTX-M-64质粒的宿主菌有大肠埃希菌(GenBank登录号KX013540、KJ020576、KP091735、AB976601、KC576517、GQ456156)、阴沟肠杆菌(GenBank登录号GQ300937)等,本研究是第一例关于宋内志贺氏菌携带完整的ISEcp1-blaCTX-M-orf477转座单元的报道。
本研究中从腹泻患者大便样本中分离到一株产ESBL的宋内志贺氏菌1173,PCR检测表明其携带2种ESBL编码基因——blaCTX-M-64和blaTEM。接合转移实验获得一个接合子1173-CTXM-EC600,也为产ESBL菌株,但其只携带blaCTX-M-64耐药基因,这说明blaCTX-M-64和blaTEM在1173基因组中的位置可能是不同的:blaCTX-M-64位于质粒上,且可通过性菌毛接合转移至受体菌EC600中,同时使接合子1173-CTXM-EC600获得了blaCTX-M-64编码的ESBL表型;blaTEM可能位于另一个不可接合质粒上或者染色体上(对后期基因组测序结果分析表明,其位于染色体上)。MIC测定结果可以看出,1173和1173-CTXM-EC600对青霉素类、单环β-内酰胺类、一代、二代和四代头孢菌素类、碳青霉烯类、喹诺酮类、呋喃类、四环素类抗菌药物的敏感性一致,都对青霉素类、单环β-内酰胺类、一代、二代和四代头孢菌素类耐药,说明1173对这些药物的耐药性是由携带blaCTX-M-64的质粒介导的,并可通过性菌毛将耐药性传递给受体菌。
| [1] | Wang L, Liu L, Liu D, Yin Z, Feng J, Zhang DF, Fang HH, Qiu YF, Chen WJ, Yang RS, Wang JL, Fa YZ, Zhou DS. The first report of a fully sequenced resistance plasmid from Shigella boydii. Frontiers in Microbiology, 2016, 7: 1579. |
| [2] |
Mahemuti, Wupuer X, Li XL, Li F, Husaiyin M, Gu BS, Zhang J, Shataer W, Tuohuti M. Analysis on groups and serotypes of Shigella in Xinjiang, 2003-2013. Chinese Journal of Preventive Medicine, 2015, 49(5): 447-449.
(in Chinese) 马合木提, 夏依旦·吾甫尔, 李新兰, 李方, 木合亚提·胡塞英, 顾本思, 张建, 外力·沙塔尔, 穆塔里普·托呼提. 2003-2013年新疆维吾尔自治区哨点医院的志贺菌菌型分布及变迁分析. 中华预防医学杂志, 2015, 49(5): 447-449. |
| [3] | Killackey SA, Sorbara MT, Girardin SE. Cellular aspects of Shigella pathogenesis:focus on the manipulation of host cell processes. Frontiers in Cellular and Infection Microbiology, 2016, 6: 38. |
| [4] | Lima IF, Havt A, Lima AA. Update on molecular epidemiology of Shigella infection. Current Opinion in Gastroenterology, 2015, 31(1): 30-37. DOI:10.1097/MOG.0000000000000136 |
| [5] | Livio S, Strockbine NA, Panchalingam S, Tennant SM, Barry EM, Marohn ME, Antonio M, Hossain A, Mandomando I, Ochieng JB, Oundo JO, Qureshi S, Ramamurthy T, Tamboura B, Adegbola RA, Hossain MJ, Saha D, Sen S, Faruque ASG, Alonso PL, Breiman RF, Zaidi AKM, Sur D, Sow SO, Berkeley LY, O'Reilly CE, Mintz ED, Biswas K, Cohen D, Farag TH, Nasrin D, Wu YK, Blackwelder WC, Kotloff KL, Nataro JP, Levine MM. Shigella isolates from the global enteric multicenter study inform vaccine development. Clinical Infectious Diseases, 2014, 59(7): 933-941. DOI:10.1093/cid/ciu468 |
| [6] | Nhu NTK, Vinh H, Nga TVT, Stabler R, Duy PT, Vien LTM, van Doorn HR, Cerdeño-Tárraga A, Thomson N, Campbell J, van Minh Hoang N, Nga TTT, van Minh P, Thuy CT, Wren B, Farrar J, Baker S. The sudden dominance of blaCTX-M harbouring plasmids in Shigella spp. Circulating in Southern Vietnam. PLoS Neglected Tropical Diseases, 2010, 4(6): e702. DOI:10.1371/journal.pntd.0000702 |
| [7] | Zhang WL, Luo YP, Li JY, Lin L, Ma Y, Hu CQ, Jin SH, Ran L, Cui SH. Wide dissemination of multidrug-resistant Shigella isolates in China. Journal of Antimicrobial Chemotherapy, 2011, 66(11): 2527-2535. DOI:10.1093/jac/dkr341 |
| [8] | Klontz KC, Singh N. Treatment of drug-resistant Shigella infections. Expert Review of Anti-Infective Therapy, 2015, 13(1): 69-80. DOI:10.1586/14787210.2015.983902 |
| [9] | Niyogi SK. Increasing antimicrobial resistance-an emerging problem in the treatment of shigellosis. Clinical Microbiology and Infection, 2007, 13(12): 1141-1143. DOI:10.1111/j.1469-0691.2007.01829.x |
| [10] | Pitout JDD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae:an emerging public-health concern. The Lancet Infectious Diseases, 2008, 8(3): 159-166. DOI:10.1016/S1473-3099(08)70041-0 |
| [11] | D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type β-lactamases:a successful story of antibiotic resistance. International Journal of Medical Microbiology, 2013, 303(6/7): 305-317. |
| [12] | Cantón R, González-Alba JM, Galán JC. CTX-M enzymes:origin and diffusion. Frontiers in Microbiology, 2012, 3: 110. |
| [13] | CLSI. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute, 2015. |
| [14] | Chen ZH, Li HX, Feng J, Li YX, Chen X, Guo XM, Chen WJ, Wang L, Lin L, Yang HY, Yang WH, Wang J, Zhou DS, Liu CT, Yin Z. NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Frontiers in Microbiology, 2015, 6: 294. |
| [15] |
Jiang XY, Liu D, Xu L, Fang HH, Feng J, Yin Z, Zhou DS, Wang L, Zhang DF, Song YJ. Resistance mechanisms of blaCTX-M-55 in a clinical Shigella sonnei strain. Military Medical Sciences, 2016, 40(9): 717-721.
(in Chinese) 蒋晓圆, 刘东, 徐丽, 方海红, 冯娇, 殷喆, 周冬生, 王丽, 张德福, 宋亚军. blaCTX-M-55介导的宋内志贺菌耐药机制研究. 军事医学, 2016, 40(9): 717-721. |
| [16] | Qu F, Ying Z, Zhang CL, Chen ZH, Chen SM, Cui EB, Bao CM, Yang HY, Wang J, Liu CT, Mao YL, Zhou DS. Plasmid-encoding extended-spectrum beta-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiology, 2014, 9(10): 1143-1150. DOI:10.2217/fmb.14.53 |
| [17] | Lü LC, Partridge SR, He LY, Zeng ZL, He DD, Ye JH, Liu JH. Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrobial Agents and Chemotherapy, 2013, 57(6): 2824-2827. DOI:10.1128/AAC.02155-12 |
| [18] | Nagano Y, Nagano N, Wachino JI, Ishikawa K, Arakawa Y. Novel chimeric β-lactamase CTX-M-64, a hybrid of CTX-M-15-like and CTX-M-14β-lactamases, found in a Shigella sonnei strain resistant to various oxyimino-cephalosporins, including ceftazidime. Antimicrobial Agents and Chemotherapy, 2009, 53(1): 69-74. DOI:10.1128/AAC.00227-08 |
| [19] | Messai Y, Ibadene H, Benhassine T, Alouache S, Tazir M, Gautier V, Arlet G, Bakour R. Prevalence and characterization of extended-spectrum β-lactamases in Klebsiella pneumoniae in Algiers hospitals (Algeria). Pathologie Biologie, 2008, 56(5): 319-325. DOI:10.1016/j.patbio.2008.05.008 |
| [20] | Tamang MD, Nam HM, Kim TS, Jang GC, Jung SC, Lim SK. Emergence of extended-spectrum β-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. Journal of Clinical Microbiology, 2011, 49(7): 2671-2675. DOI:10.1128/JCM.00754-11 |
| [21] | Ma L, Siu LK, Lu PL. Effect of spacer sequences between blaCTX-M and ISEcp1 on blaCTX-M expression. Journal of Medical Microbiology, 2011, 60(12): 1787-1792. DOI:10.1099/jmm.0.033910-0 |
 2018, Vol. 58
2018, Vol. 58