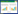中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 祁双双, 吴海珍, 叶江, 张惠展. 2018
- Shuangshuang Qi, Haizhen Wu, Jiang Ye, Huizhan Zhang. 2018
- bagZH编码酪氨酸酶样铜酶并参与bagremycin生物合成
- BagZH encodes tyrosinase-like copper enzyme and participates in bagremycin biosynthesis
- 微生物学报, 58(12): 2229-2239
- Acta Microbiologica Sinica, 58(12): 2229-2239
-
文章历史
- 收稿日期:2018-04-04
- 修回日期:2018-05-17
- 网络出版日期:2018-06-07
分离自印度尼西亚爪哇土壤中的链霉菌Tü4128株(Streptomyces sp. Tü4128)能合成bagremycin A和bagremycin B两种新型次级代谢产物,两者对芽孢杆菌属、链霉菌属、节杆菌属等一些革兰氏阳性菌具有一定的抗菌活性[1]。Bagremycin B是铁载体绿铁(ferroverdin)的化学降解物,但作为链霉菌次级代谢的天然产物还是首次被报道[2]。
在bagremycin生物合成途径中[1],合成bagremycin A的前体化合物是反式对香豆酸(p-coumaric)和3-氨基-4-羟基苯甲酸(3, 4-AHBA)。作为肉桂酸(cinnamic acid)的衍生物,反式对香豆酸来源于莽草酸途径提供的苯丙氨酸(图 1-A)。3, 4-AHBA由三碳化合物与四碳化合物缩合而成,其路线常见于手霉素和阿克苏霉素等抗生素的合成途径中[3-4]。

|
| 图 1 链霉菌Tü4128株bagremycin生物合成途径及其基因簇 Figure 1 Hypothetical biosynthesis pathway and gene cluster of bagremycin. A: Hypothetical biosynthesis pathway of bagremycin. B: The gene cluster of bagremycin. |
我们在前期工作中采用Solexa/Illumia技术对链霉菌Tü4128株进行了全基因组测序和bagremycin生物合成基因簇(bag)定位,但其上下游边界尚未确定。bag基因簇中的bagA、bagB、bagC、bagE、bagI和bagJ基因已被克隆和鉴定[5-8],其中BagB和BagC负责反式对香豆酸的合成。
bagZ与bagH是Tü4128株bag基因簇中紧邻的两个同向基因(图 1-B)。经同源比对分析,BagH属于酪氨酸酶家族的氨基酚氧化酶。酪氨酸酶是存在于所有生物体内的含铜酶[9],属于双核铜蛋白家族中的一类[10-11]。
本文证实BagH为酪氨酸酶家族的新成员,BagZ作为辅因子激活BagH酶活性。BagH重组蛋白(rBagH)在体外能以氨基酚氧化酶的作用催化o-氨基酚底物,暗示BagH在体内氧化前体化合物3, 4-AHBA的邻位氨基,由此参与bagremycin的生物合成。
1 材料和方法 1.1 菌株和质粒用于菌种保藏、接合转移和重组子筛选的链霉菌Tü4128株孢子生长在SMA培养基上,YEME培养基用于菌丝体培养,发酵培养基用于次级代谢产物的合成和分析。上述三种培养基按照前文所列的配方[5]配制。本文所涉菌种和质粒的性质分列在表 1中。
| Strains or plasmids | Characteristics | Source |
| E. coli JM83 | Fʹ, ara, lac-proAB, rpsL, (Strr), Φ80d lacZ △M15 | Laboratory stock |
| E. coli BL21 (DE3) | F–, omp, hsdSB(rB–, mB–), gal dcm(DE3) | Laboratory stock |
| E. coli ET12567 | dam-13::Tn9, dcm-6, hsdM, hsdR, recF-143, zjj-201::Tn10, galK-2, galT-22, ara-14, lacY-1, xy-l5, leuB-6, thi-1, tonA-31, rpsL-136, hisG-4, tsx-78, mtli, glnV-44, F−, Cmr | Professor Chang-Sheng Zhang |
| Streptomyces sp. Tü4128 | Wild type bagremycins producing stain | Professor Hans-Peter Fiedler |
| Tü4128-△bagZH | In-frame deletion strain of bagZH, Kmr | This study |
| Tü4128-△bagZH:bagZH | Tü4128-△bagZH complemented with bagZH, Kmr, Amr | This study |
| Tü4128-bagZH | Tü4128 complemented with bagZH, Kmr, Amr | This study |
| pMJ | oriT, Kmr, Amr | This study |
| pIB139 | aac3(IV), attφC31, oriT, Amr, PermE* | Laboratory stock |
| pET-43.1a(+) | Apr, PT7 | Laboratory stock |
| pMJ-△bagZH | Deletion plasmid of bagZH, Kmr, Amr | This study |
| pIB139-bagZH | Complementary plasmid of bagZH, Amr | This study |
| pET-43.1a-bagH | Expression plasmid of bagH, Apr | This study |
1.2 基因敲除、回补和过表达
PCR克隆bagZH基因上下游同源臂,分别连接至pMJ质粒neo基因的上游和下游,构成敲除质粒pMJ-△bagZH。将之转化至E. coli ET12567感受态细胞,DNA水平验证正确的克隆用于链霉菌Tü4128株的接合转移,借助同源重组敲除bagZH基因。由阿泊拉霉素抗性筛选出的二次重组子再经DNA水平验证,并进行RT-PCR检验以排除bagZH灭活操作本身影响上下游基因转录的可能性,最终获得链霉菌Tü4128株的bagZH基因敲除株(Streptomyces sp. Tü4128-△bagZH)。
将bagZH基因的ORF PCR克隆至链霉菌表达型质粒pIB139的PermE*启动子下游,构成pIB139-bagZH回补质粒。通过接合转移将回补质粒分别克隆至敲除株Tü4128-ΔbagZH和野生株Tü4128中,完成回补株和过表达株的构建;同样,将pIB139空载质粒克隆至敲除株和野生株,作为对照菌株。
1.3 HPLC检测次级代谢产物在28 ℃和摇床转速为180 r/min条件下发酵培养野生株(或对照株)、敲除株、回补株和过表达株。一级摇瓶(种子)培养3 d,二级摇瓶培养14 d (335 h)。发酵液经离心去除菌体,用HCl将发酵上清液调至pH 5,过滤杂质,加入等体积的乙酸乙酯萃取,旋转蒸发,用1.2 mL甲醇溶解发酵产物,过滤后进行HPLC分析。HPLC检测采用XDB-C18柱,流动相为40%乙腈与60%浓度为0.1% (V/V)磷酸的混合溶液,流速为1 mL/min,柱温30 ℃,280 nm进行DAD检测。所有样品进样20 μL。
1.4 重组蛋白表达与分离纯化PCR扩增bagH基因的ORF,采用In-fusion程序将之克隆至大肠杆菌表达型pET-43.1a(+),构成pET-43.1a-bagH重组质粒。该构建物保留了肠激酶酶切位点,便于将重组蛋白rBagH中的Nus标签切除。经验证正确的重组质粒转化E. coli BL21 (DE3)感受态细胞,进行体外诱导表达。表达后的菌体经超声破碎和离心获取上清液(粗酶液),用于重组蛋白rBagH的镍螯合亲和层析分离纯化。重组蛋白经考马斯亮蓝染色,测定595 nm处吸光值,最终根据蛋白质标准曲线定量重组蛋白的浓度。
将纯化后的重组蛋白溶液转移至透析袋中,用1×重组牛肠激酶反应缓冲液(50 mmol/L Tris-HCl,pH 8.0,1 mmol/L CaCl2,0.1% Tween-20)在4 ℃下进行透析。透析缓冲液与蛋白溶液的体积比大于20:1。透析后的重组蛋白于16 ℃酶切12 h,切除Nus标签。
1.5 BagH蛋白体外活性分析rBagH催化邻氨基酚氧化的活性分析总体系为1 mL,其中1.5 mmol/L邻氨基酚底物,50 mmol/L磷酸钾(pH 7.4),重组蛋白BagH (3.3 μg– 330 μg)。所有实验组均设置不添加酶的对照组。具体反应流程为:未加酶的反应液在30 ℃预孵育10 min;加酶(rBagH)后反应液在30 ℃反应30 min;加入100 μL 5% (V/V)三氟乙酸终止反应,反应产物过滤后进行HPLC分析。检测使用的XDB-C18柱经0.1% (V/V)三氟乙酸平衡3 min,再用线性梯度为0–100% (V/V)的含0.1% (V/V)三氟乙酸的乙腈洗脱15 min,流速为1 mL/min,270 nm进行DAD检测[12]。所有样品进样20 μL。
2 结果 2.1 bagH和bagZ的生物信息学分析bagH基因全长924 bp,编码308个氨基酸,蛋白分子量约为34 kDa。BagH的系统发育树见图 2。在NCBI数据库中进行同源比对,BagH与链霉菌Streptomyces murayamaensis中的NspF[13-14]一致性达59%,预测为铜蛋白家族Ⅲ型酪氨酸酶超家族成员邻氨基酚氧化酶。该超家族成员均需铜离子激活,每个酶分子结合2个铜离子[15-16]。
|
| 图 2 BagH系统发育树 Figure 2 Phylogenetic analysis of BagH and their homologs. Sequence alignments were performed with ClustalW, and the neighbor-joining tree was generated with Mega 6. Numbers on the tree branches represent the bootstrap support calculated per 1000 bootstrap replicates. BagH is indicated by bold. |
bagZ基因全长357 bp,编码119个氨基酸,蛋白分子量约为13 kDa。在NCBI数据库中进行同源比对,BagZ与MelC1一致性高达97%, 与Streptomyces griseus中grixazone生物合成途径的GriE一致性达55%,与Streptomyces murayamaensis中4, 3-HNBAm生物合成途径的NspE一致性达47%。MelC1是MelC2的分子伴侣,协助MelC2多肽链正确折叠,同时负责将Cu2+转运至MelC2的活性中心[17-18]。MelC2是典型的酪氨酸酶,在多种链霉菌中负责次级代谢产物黑色素的合成[18]。鉴于BagH与MelC2同源比对的一致性高达61%,预示BagZ与MelC1发挥相似的功能,负责BagH多肽链折叠与Cu2+转运。
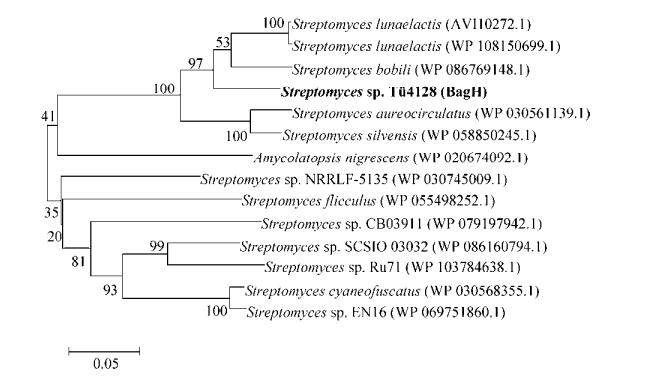
2.2 敲除株Tü4128-ΔbagZH的构建分别PCR克隆bagZH基因上游同源臂(621 bp)的下游同源臂(793 bp),并按图 3-A所示程序构建敲除质粒pMJ-△bagZH。含pMJ-△bagZH的E. coli ET12567与链霉菌Tü4128株接合转移,覆盖抗生素筛选二次重组子(图 3-B)。
|
| 图 3 链霉菌Tü4128-ΔbagZH敲除株的构建与筛选 Figure 3 Construction and verification of bagZH disruption mutant. A: The schematic diagram of disruption plasmids. B: Resistant screening of target recombinants. Left: kanamycin; right: apramycin. C: Transcripts in Streptomyces sp. Tü4128. M: DNA marker; Lanes 1, 2: Negative control, primers of bagG; 3: bagG (238 bp); 4-6: Positive control, primers of bagA, bagC and bagE; 7, 8: Negative control, primers of bagI; 9: bagI (251 bp). D: Transcripts in Streptomyces sp. Tü4128-ΔbagZH. M: DNA marker; Lanes 1, 2: Negative control, primers of bagG; lane 3: bagG (238 bp); lane 4-6: Positive control, primers of bagA, bagC and bagE; lane 7, 8: Negative control, primers of bagI; lane 9: bagI (251 bp). |
为了排除bagZH基因敲除操作本身影响上下游基因正常表达的可能性,分别从培养11 d的野生株Tü4128和敲除株Tü4128-ΔbagZH中提取总RNA,经DNaseⅠ消化后逆转录为cDNA。PCR检测bagZH上游基因bagG和下游基因bagI的转录水平,并比较野生株(图 3-C)与敲除株(图 3-D)的RT-PCR格局。
结果显示,无论野生株Tü4128还是敲除株Tü4128-ΔbagZH,3个阳性对照基因正常转录,bagZH上下游基因bagG和bagI的转录也没有发生显著改变,而所有阴性对照在对应大小处均无扩增产物。可见,bagZH基因的敲除操作并不影响其上下游基因的转录,由ΔbagZH造成的次级代谢产物谱改变可视为bagZH基因的贡献。
2.3 bagZH与bagremycin生物合成的相关性为了探查bagZH灭活对bagremycin合成的影响,将野生株Tü4128、敲除株Tü4128-ΔbagZH、回补株Tü4128-△bagZH:bagZH和过表达株Tü4128:bagZH分别发酵培养14 d (335 h),HPLC检测四株菌的bagremycin A和B水平(图 4-B和4-C)。Bagremycin A和bagremycin B的质谱HPLC/ TOF-MS分析结果显示,在本文所述的检测条件下,bagremycin A和B两组分的保留时间分别为7.8 min和16.7 min (图 4-A)。

|
| 图 4 Streptomyces sp. Tü4128与敲除株、回补株、过表达株的发酵产物HPLC结果 Figure 4 HPLC analysis of bagremycins in bagZH mutant strains, complementation strains and overexpression strains. A: HPLC analysis and retention time of bagremycin A and B. B: Production of bagremycin A. C: Production of bagremycin B. a Values were expressed as the mean±standard deviation obtained from three independent experiments. b mAU*s was defined as the amount of peak area that accumulated the amount of production at a rate of peak height per second. |
发酵结果和bagremycin的HPLC峰面积统计分析显示,bagZH联合敲除后,bagremycin A和B组分的产量均呈显著下降;在敲除株中回补bagZH双基因后,bagremycin的两种组分产量均有所提高,但没有恢复到野生株水平;若在野生株中导入bagZH双基因表达盒(即过表达株),bagremycin A和B组分的产量高于野生株。上述结果证实bagZH基因影响bagremycins的生物合成。
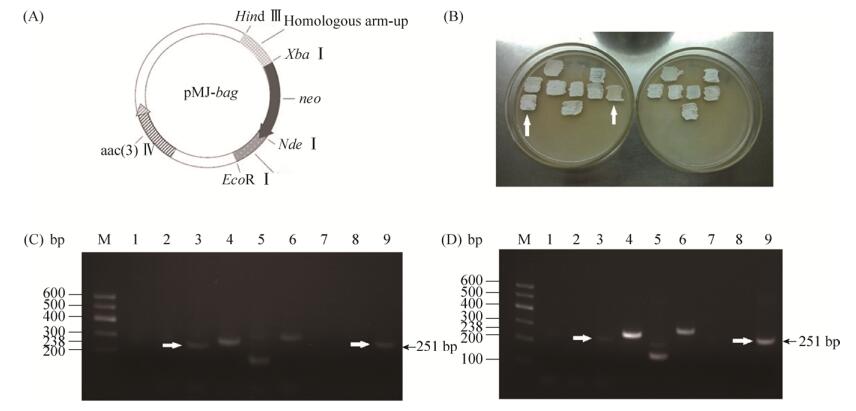
2.4 野生株Tü4128和敲除株Tü4128-ΔbagZH的次级代谢产物谱多批次发酵产物的HPLC结果显示(图 5-A),相比野生株Tü4128,在敲除株Tü4128-ΔbagZH的次级代谢产物谱中,保留时间为3.00 min和6.31 min的产物不再合成,而在保留时间为3.18 min处出现的新产物积累。推测bagZH基因敲除后,失去保护基团的邻位氨基与另一3, 4-AHBA分子的羟基发生酯化反应,产生分子量为285.24 g/mol的化合物C14H9O5N2。敲除株Tü4128-ΔbagZH的LC-ESI-MS结果显示,保留时间为3.18 min的新产物分子量为286.32 g/mol (图 5-B和5-C)。
|
| 图 5 野生株Tü4128和敲除株Tü4128-ΔbagZH的次级代谢产物谱 Figure 5 Secondary metabolite spectrum of Streptomyces sp. Tü4128 and Tü4128-ΔbagZH (A), LC-ESI-MS analysis of Tü4128 (B) and Tü4128-ΔbagZH (C). |
2.5 重组BagH的体外生化分析
重组质粒pET-43.1a-bagH在大肠杆菌BL21 (DE3)中几乎以完全可溶形式表达(图 6-A),但含重组质粒pET-43.1a-bagZ的大肠杆菌BL21 (DE3)经诱导后却以包涵体形式表达重组蛋白rBagZ (图 6-B)。为了获得具有活性的单体酶,我们在rBagH粗酶液中加入终浓度为1 mmol/L的CuSO4 (代替BagZ转运Cu2+的作用,BagH与Cu2+的摩尔比为1:2)。经Ni2+-NTA层析纯化后的rBagH约为95.7 kDa (图 6-A),肠激酶切除Nus标签,获得可用于酶活检测的纯酶液。

|
| 图 6 SDS-PAGE检测重组蛋白BagH与BagZ表达 Figure 6 SDS-PAGE of recombinant protein BagH and BagZ. M: Protein marker. (A) lane 1: Inclusion body of pET43.1a-bagH; lane 2: Supernatant of pET43.1a-bagH. (B) lane 1: Inclusion body of pET43.1a-bagZ; lane 2: Supernatant of pET43.1a-bagZ. |
鉴于生物信息学预测BagH是以邻氨基酚为底物的氧化酶(将邻位氨基氧化成亚硝基),我们推测在bagremycin生物合成途径中,BagH可能负责将3-氨基-4-羟基苯甲酸(3, 4-AHBA,邻氨基酚结构类似物)中的氨基氧化为亚硝基。因此,我们选择3, 4-AHBA和邻氨基酚作为底物(图 7-A),HPLC检测分析BagH对两种底物的体外活性。
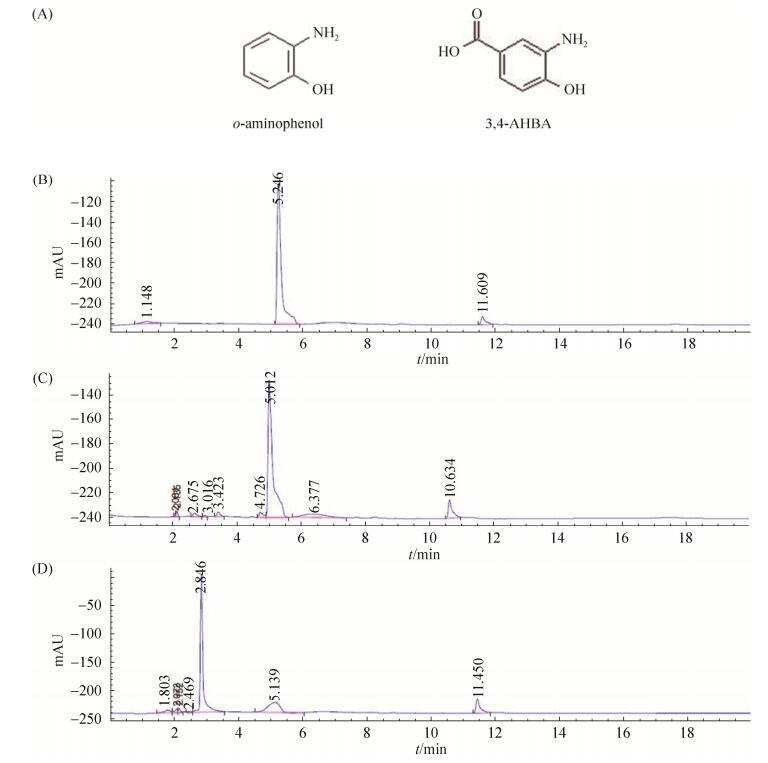
|
| 图 7 邻氨基酚与3-氨基-4-羟基苯甲酸的化学结构和邻氨基酚体外活性分析 Figure 7 A: Chemical structure of o-aminophenol and 3, 4-AHBA and in vitro activity analysis of o-aminophenol. B: o-aminophenol. C: Control group, adding proteins of pET-43.1a (+). D: Experimental group, adding rBagH. |
HPLC结果显示(图 7-C和7-D),检测波长为270 nm时,实验组中保留时间为5.2 min的邻氨基酚底物峰较之对照组减少,同时产生新的产物(保留时间2.8 min)。表明BagH能将邻氨基酚催化产生新的产物(其结构未鉴定)。然而,在同样条件下实验组中保留时间为4.3 min的底物3, 4-AHBA并没有减少,也没有新的产物产生。推测在体外环境下,BagH对酚式羟基的对位基团具有选择性,基团越简单,其邻位氨基就越容易被氧化。支持这一推测的事实是:BagH的同源蛋白rNspF在氧化底物亚硝基化时[19],底物酚式羟基的对位基团会对rNspF的活性产生很大影响;rNspF可以氧化3, 4-AHBA,但对3, 4-AHBAm的活性更高[12]。
3 讨论本文调查结果显示,联合行使邻氨基酚氧化酶活性的链霉菌Tü4128株BagZH参与bagremycin的生物合成,但似乎并非bagremycin生物合成所必需。我们在Tü4128全基因测序分析时发现(另文待发表),Tü4128株基因组中存在包括bag在内的多种次级代谢产物生物合成基因簇,其中不乏BagH的同工酶基因。bagZH基因被敲除后bagremycin产量降低,但没有完全消失,意味着Tü4128株细胞内存在多重代谢旁路发挥BagH样的功能。
HPLC结果显示,BagH可在体外以邻氨基酚为底物催化产生新的产物,推测BagH将邻氨基酚苯环上的氨基氧化为亚硝基(图 5-B和5-C)。然而,在相同反应条件下,3, 4-AHBA未能被BagH氧化,这可能是由于体外测活体系与体内环境的不同所致。由于我们未能实现bagZ与bagH基因共表达,因而构建了pET-43.1a-bagH重组质粒将bagH单独体外表达,并在超声破碎时加入Cu2+代替BagZ的作用。据文献报道,纯化时为避免NspE(BagZ同源伙伴)的干扰单独表达NspF,加入足够高浓度的Cu2+可以代替NspE激活NspF的酶活性[12]。我们使用文献推荐的Cu2+浓度却未能将3, 4-AHBA的邻位氨基氧化,有可能该浓度对于BagH体外氧化3, 4-AHBA仍不够高。另外,在体外环境下,BagH对邻氨基酚类底物酚式羟基对位基团的偏爱性也是一种可能的解释。
综上所述,本文首次鉴定了bagZH基因与bagremycin生物合成的相关性,即bagZH以编码酪氨酸酶样铜酶的方式参与bagremycin生物合成。作为酪氨酸酶家族的一员,BagH在链霉菌Tü4128株的bagremycin生物合成途径中,可能贡献于3, 4-AHBA邻位氨基的氧化保护,避免3, 4-AHBA在体内发生自身酯化,从而促进3, 4-AHBA与反式对香豆酸酯化产生bagremycin前体化合物。据报道,作为抗生素的bagremycin对特定类型的腺癌也有一定的抑制作用[1, 20],因而我们的研究结果为bagremycin作用机制的深入研究以及高产菌株的理性设计与改造提供了基础和参考。
| [1] | Jonas R, Pandey A, Tharun G. Biotechnological advances and applications in bioconversion of renewable raw materials. Braunschweig, Germany: Doehring Druck, 2004. |
| [2] | Ballio A, Bertholdt H, Carilli A, Chain EB, di Vittorio V, Tonolo A, Vero-Barcellona L. Studies on ferroverdin, a green iron-containing pigment produced by a Streptomyces Wak. species. Proceedings of the Royal Society B: Biological Sciences, 1963, 158(970): 43-70. DOI:10.1098/rspb.1963.0033 |
| [3] | Floss HG. Natural products derived from unusual variants of the shikimate pathway. Natural Product Reports, 1997, 14(5): 433-452. DOI:10.1039/np9971400433 |
| [4] | Li YF, Gould SJ, Proteau PJ. Biosynthesis of 3-amino-4-hydroxybenzoic acid in Streptomyces murayamaensis: incorporation of[4-13C] oxalacetate. Tetrahedron Letters, 2000, 41(27): 5181-5185. DOI:10.1016/S0040-4039(00)00772-3 |
| [5] | Zhu YX, Xu DK, Liao SY, Ye J, Zhang HZ. Cloning and characterization of bagB and bagC, two co-transcribed genes involved in bagremycin biosynthesis in Streptomyces sp. Tü 4128. Annals of Microbiology, 2013, 63(1): 167-172. |
| [6] | Zhu YX, Liao SY, Ye J, Zhang HZ. Cloning and characterization of a novel tyrosine ammonia lyase-encoding gene involved in bagremycins biosynthesis in Streptomyces sp. Biotechnology Letters, 2012, 34(2): 269-274. |
| [7] | Liu F, Xu DK, Zhang YC, Zhu YX, Ye J, Zhang HZ. Identification of BagI as a positive transcriptional regulator of bagremycin biosynthesis in engineered Streptomyces sp. Tü4128. Microbiological Research, 2015, 173: 18-24. DOI:10.1016/j.micres.2015.01.011 |
| [8] |
Zhang YC, Wu HZ, Ju C, Qi SS, Ye J, Zhang HZ. Identification of bagJ as a resistant gene for novel antibiotic bagremycins in Streptomyces sp. Tü4128. Journal of East China University of Science and Technology (Natural Science Edition), 2017, 43(2): 184-192.
(in Chinese) 张玉琛, 吴海珍, 鞠诚, 祁双双, 叶江, 张惠展. Streptomyces sp. Tü4128中新型抗生素bagremycins抗性基因bagJ的研究. 华东理工大学学报(自然科学版), 2017, 43(2): 184-192. |
| [9] | Claus H, Decker H. Bacterial tyrosinases. Systematic and Applied Microbiology, 2006, 29(1): 3-14. DOI:10.1016/j.syapm.2005.07.012 |
| [10] | Faccio G, Kruus K, Saloheimo M, Th ny-Meyer L. Bacterial tyrosinases and their applications. Process Biochemistry, 2012, 47(12): 1749-1760. DOI:10.1016/j.procbio.2012.08.018 |
| [11] | Ramsden CA, Riley PA. Tyrosinase: the four oxidation states of the active site and their relevance to enzymatic activation, oxidation and inactivation. Bioorganic & Medicinal Chemistry, 2014, 22(8): 2388-2395. |
| [12] | Noguchi A, Kitamura T, Onaka H, Horinouchi H, Ohnishi Y. A copper-containing oxidase catalyzes C-nitrosation in nitrosobenzamide biosynthesis. Nature Chemical Biology, 2010, 6(9): 641-643. DOI:10.1038/nchembio.418 |
| [13] | Ginsbach JW, Kieber-Emmons MT, Nomoto R, Noguchi A, Ohnishi Y, Solomon EI. Structure/function correlations among coupled binuclear copper proteins through spectroscopic and reactivity studies of NspF. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(27): 10793-10797. DOI:10.1073/pnas.1208718109 |
| [14] | Cone MC, Melville CR, Carney JR, Gore MP, Gould SJ. 4-hydroxy-3-nitrosobenzamide and its ferrous chelate from Streptomyces murayamaensis. Tetrahedron, 1995, 51(11): 3095-3102. DOI:10.1016/0040-4020(95)00071-F |
| [15] | Tepper AWJW, Bubacco L, Canters GW. Interaction between the type-3 copper protein tyrosinase and the substrate analogue p-nitrophenol studied by NMR. Journal of the American Chemical Society, 2005, 127(2): 567-575. DOI:10.1021/ja0454687 |
| [16] | Worrall JAR, Vijgenboom E. Copper mining in Streptomyces: enzymes, natural products and development. Natural Product Reports, 2010, 27(5): 742-756. DOI:10.1039/b804465c |
| [17] | Chen LY, Chen MY, Leu WM, Tsai TY, Lee YH. Mutational study of Streptomyces tyrosinase trans-activator MelC1. MelC1 is likely a chaperone for apotyrosinase. Journal of Biological Chemistry, 1993, 268(25): 18710-18716. |
| [18] | Leu WM, Chen LY, Liaw LL, Lee YH. Secretion of the Streptomyces tyrosinase is mediated through its trans-activator protein, MelC1. Journal of Biological Chemistry, 1992, 267(28): 20108-20113. |
| [19] | Noguchi A, Horinouchi S, Ohnishi Y. Substrate specificity of benzamide synthetase involved in 4-hydroxy-3-nitrosobenzamide biosynthesis. The Journal of Antibiotics, 2011, 64(1): 93-96. |
| [20] | Bertasso M, Holzenk mpfer M, Zeeck A, Dall'Antonia F, Fiedler HP. Bagremycin A and B, novel antibiotics from Streptomyces sp. Tü4128. Journal of Antibiotics, 2001, 54(9): 730-736. DOI:10.7164/antibiotics.54.730 |
 2018, Vol. 58
2018, Vol. 58