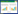中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 胡悦, 蔡英丽, 喻晶晶, 周雁, 李军平, 边银丙, 龚钰华. 2018
- Yue Hu, Yingli Cai, Jingjing Yu, Yan Zhou, Junping Li, Yinbing Bian, Yuhua Gong. 2018
- 香菇HMG-box转录因子lelcrp1基因的功能
- Function analysis of HMG-box transcription factor lelcrp1 in Lentinula edodes
- 微生物学报, 58(12): 2100-2109
- Acta Microbiologica Sinica, 58(12): 2100-2109
-
文章历史
- 收稿日期:2017-12-26
- 修回日期:2018-01-29
- 网络出版日期:2018-03-21
香菇(Lentinula edodes)是我国主要的人工栽培食用菌之一,目前主要采用栎木屑为栽培原料,但栽培料中干物质转化利用率较低,且尚未普遍采用秸秆作为栽培原料[1]。深入研究香菇对栽培基质中木质纤维素降解的分子机制,对提高栽培基质转化利用率,拓展栽培基质种类均具有重要的意义。
香菇作为一种典型的白腐真菌,主要通过分泌到胞外的木质纤维素酶降解纤维素。真菌木质纤维素酶活与相应酶基因的表达具有一致性,提高真菌胞外木质纤维素相关酶基因表达水平有利于促进栽培基质有效利用。真菌木质纤维素酶基因的表达受多种转录因子调控,目前已鉴定的参与调控纤维素酶基因、半纤维素酶基因转录的正调控因子有Xyr1、Ace2、Hap2/3/5、ClrB,负调控因子有Ace1、Cre1,这些转录因子主要存在于丝状真菌中[2]。在白腐真菌中仅有少数与木质纤维素酶基因表达相关的转录因子已经被报道,但功能尚不明确[3]。
高迁移率蛋白家族(HMG)是染色质的组成蛋白,通常具有1-7个HMG-box结构域,具有单个结构域的蛋白可与特异性DNA结合,而具有多个HMG-box结构域的蛋白则表现出与DNA的非特异性结合[4]。在已报道的真菌中,结合非特异性DNA的HMG蛋白包括酿酒酵母(Saccharomyces cerevisiae) Hmo1p和Hmo2p[5-6],Abf2p[7]和Nhp6Ap/Nhp6Bp[8],粟酒裂殖酵母(Schizosaccharomyces pombe) Hmo1[9]及柄孢霉(Podospora anserine) mtHMG1[10]。Ohm等研究发现,裂褶菌(Schizophyllum commune)具有HMG-box结构域的转录因子hom1和hom2,与菌丝生长及子实体形成具有重要关系,敲除hom1后可以形成数量较多但个体较小的子实体,敲除hom2后则子实体不能发育;此外,裂褶菌野生型菌落形态为非对称型,而敲除hom2后菌落形态变为对称型,表明hom2对裂褶菌菌落形态也有明显的调控作用[11]。施乐乐等对金针菇(Flammulina velutipes) fvhom1基因进行过表达后,发现菌丝生长速度明显减缓[12]。由此可见,这类含有HMG-box结构域的转录因子具有调控真菌菌丝生长和子实体形成的重要功能,但关于其调控木质纤维素降解相关酶基因表达的研究却鲜有报道。
前期通过以纤维素、纤维素+木质素磺酸钠及葡萄糖为主要碳源培养的香菇菌丝转录组(数据暂未公开)和分泌蛋白组[13]比较分析,发现CAZy酶家族GH3、GH5、GH6、GH7、GH10、GH12、AA9及木质素降解酶相关部分基因或蛋白受纤维素+木质素磺酸钠诱导表达,且与香菇HMG-box型转录因子lelcrp1 (Lentinula edodes lignocellulase genes regulation protein)表达模式一致。在此基础上,本文通过RNAi方法进行lelcrp1基因沉默,并通过检测香菇与木质纤维素降解相关酶基因的表达水平变化,分析lelcrp1的基因功能。
1 材料和方法 1.1 材料 1.1.1 供试菌株:香菇异核体菌株“武香一号” W1 (ACCC 50926)、载体质粒pCAMBIA1300-g均由本实验室保藏并提供;大肠杆菌菌株DH5α、农杆菌菌株AGL1均购自上海唯地生物技术有限公司。
1.1.2 培养基及试剂:MM培养基、IM培养基和Co-IM培养基配方参考[14],LB和MYG等常规培养基配方参考[15]。Phanta Super-Fidelity DNA Polymerase、ClonExpress Multis One Step Cloning Kit、HiScript Ⅱ One Step RT-PCR Kit和AceQTM Qpcr SYBR Green Master Mix购自Vazyme公司,限制性内切酶KpnⅠ、EcoR Ⅰ、BamHⅠ和XhoⅠ购自Thermo Scientific公司,San Prep柱式DNA回收试剂盒和真菌基因组DNA快速抽提试剂盒购自生工生物工程(上海)股份有限公司,RNAiso Plus购自TaKaRa公司。
1.2 LEHMO1蛋白结构分析基于本实验室香菇单核体基因组测序[16]数据,通过网站https://www.ncbi.nlm.nih.gov/S-tructure/cdd/wrpsb.cgi对LELCRP1蛋白的结构域进行分析,蛋白结构图使用IBS软件进行绘制[17]。
1.3 lelcrp1基因沉默载体构建以W1单核体DNA及异核体cDNA为模板,分别对GPD启动子和lelcrp1基因antisense片段进行扩增,用限制性内切酶KpnⅠ和EcoRⅠ对质粒pCAMBIA1300-g进行双酶切,回收扩增片段和酶切大片段,通过double-joint[18]和同源重组构建lelcrp1基因沉默载体pCAMBIA1300-g-lelcrp1。
1.4 遗传转化体系构建及转化子筛选将含有重组质粒pCAMBIA1300-g-lelcrp1的农杆菌AGL1划线培养1 d,挑取单菌落于1 mL LB (50 μg/mL Rif+, 50 μg/mL Kan+)培养液中,200 r/min 28 ℃培养1 d。取1 mL农杆菌菌液加入到100 mL MM (50 μg/mL Kan+)液体培养基,200 r/min 28 ℃培养2 d。收集菌液,5000 r/min离心10 min,去上清后,用等体积IM培养液重悬,并用IM稀释至OD600=0.4,添加乙酰丁香酮(AS)至终浓度为200 μmol/L,于200 r/min 28 ℃培养9 h至OD600=0.6。用打孔器打取直径相同的已活化W1菌块浸入上述IM农杆菌菌液,20 min后转入含有200 μmol/L AS的Co-IM培养基,25 ℃正置培养,3 d后用无菌水冲洗菌丝3次,浸入含有400 μg/mL头孢噻肟霉素的无菌水中,20 min后转入含有3.5 μg/mL潮霉素和400 μg/mL头孢噻肟霉素的MYG培养基上,25 ℃培养。菌丝萌发后,在含有3.5 μg/mL潮霉素MYG平板上对转化子进行筛选,利用微波法[19]提取菌丝总DNA,通过启动子与antisense片段的扩增和实时荧光定量PCR检测lelcrp1基因表达水平鉴定阳性转化子。PCR上游引物为gpd-ys-F:5′- TTGCCTCTAATCCCTT GCTA-3′,下游引物为lelcrp1-anti-R:5′-CGAGGCT CCGCTCACTGTTT-3,荧光定量PCR引物为lelcrp1-RT-F和lelcrp1-RT-R (表 1)。
| Primer name | Primer sequence (5′→3′) |
| lelcrp1-RT-F | CAAGGACGGGAAGGCGAGAA |
| lelcrp1-RT-R | GAGGAGCGACGACGAACGAC |
| LE01Gene04541-RT-F | CTGCCCCCACCGTTGCATCAGCCCT |
| LE01Gene04541-RT-R | TCAAAGTAGACGACTGATTACCCAC |
| LE01Gene08136-RT-F | TTCCGTGTTACGTTCTTGATGG |
| LE01Gene08136-RT-R | CACTATTGGGCTGTGAGGATGG |
| LE01Gene12864-RT-F | GGCTTGAACGGTGCCCTTTATT |
| LE01Gene12864-RT-R | TGGACATTGGGAGTCGCAGTAG |
| LE01Gene04089-RT-F | TGGTCCTCGTTCTCAGTATTTGG |
| LE01Gene04089-RT-R | GCTGGAGGTAGTAGTGCCCGTA |
| LE01Gene04829-RT-F | TCTGGGACGATTACGCTGTGAA |
| LE01Gene04829-RT-R | CGTGGATGTGGCAGAGGAAGTA |
| LE01Gene07961-RT-F | TTTCCGCCATTCGTCGGTTGTA |
| LE01Gene07961-RT-R | GTGGTTCCGTCAGTCGGGTAGG |
| LE01Gene10050-RT-F | AAGGTGATCCCAACTACGACGAG |
| LE01Gene10050-RT-R | TACCCAAACGATAGAATCAATCAAGG |
| LE01Gene10512-RT-F | CTTCAGGCTCCGTCTCCTTTAT |
| LE01Gene10512-RT-R | ATCCCAGATGCTCATTTCTTTCAG |
| LE01Gene10634-RT-F | CTGCCGTCGTATGGAGTGGTT |
| LE01Gene10634-RT-R | TGTGCGGAATAATCGGTTGTG |
| LE01Gene07574-RT-F | TGATTTGGTTCTCGCTGTTGC |
| LE01Gene07574-RT-R | CGTCCTGAGGGATTCCATTCG |
| LE01Gene10266-RT-F | TCAACTCCAAGCGGACAATACC |
| LE01Gene10266-RT-R | CAGAATAAGCACCTGGGAAAGC |
| LE01Gene09248-RT-F | TCCCATTCAGGATGCCACAG |
| LE01Gene09248-RT-R | TTGATTCCAGTACGCGGTGA |
| LE01Gene11039-RT-F | TCTCACTGGCTCCGCTTCAACCT |
| LE01Gene11039-RT-R | CAGCCGCTTCCGCCTCTTTCATA |
| LE01Gene12502-RT-F | TACTTATGTTGCGGACCCTTCTT |
| LE01Gene12502-RT-R | AGCCTTGTTCCAAACCGAGTGA |
| LE01Gene13053-RT-F | ACCGAGTTCATCGCACAGCAAG |
| LE01Gene13053-RT-R | GTTAGGTTCGTTATCAAAGGCAAA |
| LE01Gene03223-RT-F | CTCGCACAAGGCAACTCACA |
| LE01Gene03223-RT-R | ATCTCCAGGTTCCATCGTCA |
| LE01Gene07975-RT-F | GATTCATTCAGTATTTCGCCTTGC |
| LE01Gene07975-RT-R | TCCCGAGTATGCTGGTTTCTTT |
| LE01Gene07762-RT-F LE01Gene07762-RT-R |
CTGGAGCCAACAACTATTACGC TCGGGAACAGCTACACTATCACTT |
| LE01Gene00726-RT-F | CGCAATGAGCAATGGAAGAAGAC |
| LE01Gene00726-RT-R | CAGCGAATGAACAATGGACATACAA |
| LE01Gene05156-RT-F | GCACAATTACACCGACACGC |
| LE01Gene05156-RT-R | AAGTATCCGAGCCGAACGAG |
| LE01Gene09060-RT-F | CTGGACGACCGAATGCTACTGC |
| LE01Gene09060-RT-R | CTGCGAGTCGAAAGTGAAGGGA |
| LE01Gene08997-RT-F | CAGCGTGTTCTGATGTCTTGATG |
| LE01Gene08997-RT-R | AGGAAATGACGAGGAAGCGAAT |
| LE01Gene04737-RT-F | ATCCGACCAGGAACTTGAACCA |
| LE01Gene04737-RT-R | GCAGTAACCACGAGATTGAGGG |
| LE01Gene04660-RT-F | AACACTGAGAATCCCATAAAGCG |
| LE01Gene04660-RT-R | GCCATCACAGTAGCGAGACCAG |
| LE01Gene04648-RT-F | GCATCCCTTCCACCTCCAT |
| LE01Gene04648-RT-R | CGGGCACAACTCAGTCCAG |
| LE01Gene02092-RT-F | GAGGAACTCACGAGGAAACACC |
| LE01Gene02092-RT-R | TGATTATTGGCTACGATGAACAC |
1.5 荧光定量PCR检测
将转化子接种于铺有一层无菌玻璃纸并含有3.5 μg/mL潮霉素的MYG平板上,出发菌株W1接种于铺有一层无菌玻璃纸的MYG平板,分别置于25 ℃恒温培养,10 d后,收集菌丝立即置于液氮冷却的研钵中进行研磨,研磨后的菌丝粉末立即进行RNA提取,未使用的菌丝粉末样品-80 ℃保存待用。样品总RNA提取使用Trizol plus (TaKaRa, Japan)试剂方法,cDNA的合成采用HiScript Ⅱ One Step RT-PCR Kit (Vazyme, China)试剂盒。根据目标基因在香菇基因组数据库中的CDS序列设计合成荧光定量PCR引物(表 1)。实时荧光定量PCR采用AceQTM qPCR SYBR Green Master Mix (TaKaRa, Japan)在BIO-RAD CFX Connect Real-Time System (USA)上按照Vazyme的qPCR实验操作手册进行,以leactin为内参基因,W1 cDNA为对照。
1.6 菌丝长速测定将转化子接种于含有3.5 μg/mL潮霉素的MYG平板上,每一菌株平板接种菌块设5个重复,置于25 ℃恒温培养。采用十字交叉法[20]记录菌丝生长速度:待菌丝开始萌发,以菌块中心为交点划十字并测量菌丝尖端直径r,待长速最快的菌丝即将长满平板时,测定菌落直径R,记菌丝生长天数为n,计算生长速率。菌丝日平均生长速度按照公式(1)计算。
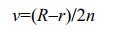
|
公式(1) |
将转化子接种于含有3.5 μg/mL潮霉素的MYG平板上,置于25 ℃恒温培养,采用真菌基因组DNA快速抽提试剂盒(生工生物工程,上海)提取高纯度DNA,用限制性内切酶BamH Ⅰ对基因组DNA进行酶切,以hpt557-F (5′-ACACTA CATGGCGTGATTTCAT-3′)和hpt557-R (5′-TC CACTATCGGCGAGTACTTCT-3′)为引物扩增hyg基因片段作为探针标记,通过酶切产物和探针的杂交结果,判断外源基因片段在香菇基因组中拷贝数[21]。
2 结果和分析 2.1 LELCRP1蛋白结构分析以香菇单核体全基因组测序数据为基础,将LELCRP1蛋白序列提交至NCBI Conserved Domain Search,分析LELCRP1蛋白的结构域组成(图 1);LELCRP1蛋白包含532个AA,有1个Ashwin super family结构域(23-113)和1个HMG-box super family结构域(124-206),后者具有HMG-box型转录因子典型的DNA-binding区域。
|
| 图 1 LELCRP1蛋白功能结构域 Figure 1 Functional motif of LELCRP1. |
2.2 基因沉默载体构建
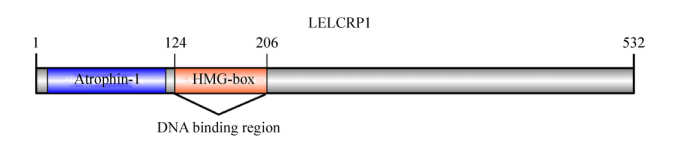
以pCAMBIA1300质粒为载体骨架,通过double-joint法连接GPD启动子(1017 bp)及lelcrp1基因antisense片段(483 bp),利用同源重组法连接片段与线性化载体,转化E. coli DH5α,通过菌落PCR选择阳性克隆子(图 2-A),同时提取质粒用限制性内切酶XhoⅠ进行酶切电泳(图 2-B)和扩增片段回收测序(P2通用引物反向测序),检测载体构建的正确性。菌落PCR、质粒酶切以及测序结果均表明构建lelcrp1基因的沉默载体成功。
|
| 图 2 重组质粒pCAMBIA1300-g-lelcrp1的菌落PCR鉴定(A)和酶切鉴定(B) Figure 2 Amplification (A) and digestion (B) of pCAMBIA1300-g-lelcrp1 cloning. A: M: DL2000 marker; lane 1: amplification of pCAMBIA1300-g- lelcrp1 cloning. B: M: 1 kb DNA ladder; lane 1: pCAMBIA1300-g-lelcrp1 cloning digested by XhoⅠ. |
2.3 菌丝转化及转化子筛选
在含有3.5 μg/mL潮霉素的MYG平板上经过4次筛选,得到了30个较为稳定的抗性转化子,微波法提取菌丝基因组总DNA。以gpd-ys-F (GPD启动子717 bp)和lelcrp1-anti-R (antisense片段483 bp)为引物对其进行特异性扩增(图 3-A),PCR产物经凝胶电泳检测,确认得到大小约为1200 bp的GPD启动子与lelcrp1基因antisense相连的片段,可以初步判定这些转化子为阳性转化子。
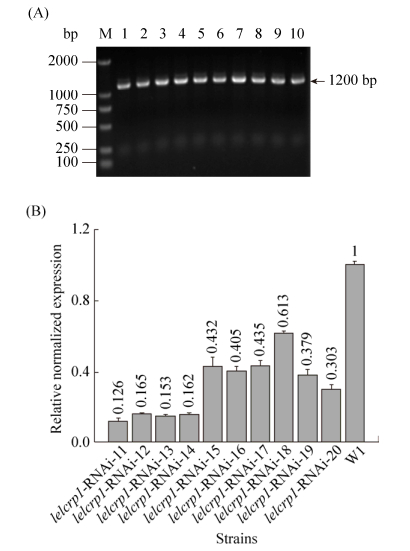
|
| 图 3 lelcrp1基因沉默转化子PCR鉴定(A)和荧光定量PCR鉴定(B) Figure 3 PCR identification (A) and fluorescence quantitative PCR identification (B) of lelcrp1 silencing transformants. A: M: DL2000 marker; lane 1-10: Transformants lelcrp1-RNAi-11, lelcrp1-RNAi-12, lelcrp1-RNAi-13, lelcrp1-RNAi-14, lelcrp1-RNAi-15, lelcrp1-RNAi-16, lelcrp1-RNAi-17, lelcrp1-RNAi-18, lelcrp1-RNAi-19, lelcrp1-RNAi-20. B: Target strains: lelcrp1-RNAi-11-20; CK: W1. |
随机挑选其中10个转化子,在含有3.5 μg/mL潮霉素的MYG玻璃纸平板上25 ℃避光培养10 d后,提取总RNA,反转录为cDNA,以lelcrp1-RT-F和lelcrp1-RT-R为引物,采用荧光定量PCR技术检测lelcrp1基因在出发菌株W1以及转化子中的表达情况(图 3-B)。结果显示,4个转化子lelcrp1-RNAi-11、lelcrp1-RNAi-12、lelcrp1-RNAi-13和lelcrp1-RNAi-14中lelcrp1基因的表达量与出发菌株相比分别下降至出发菌株的12.6%、16.5%、15.3%和16.2%,表明lelcrp1基因表达在这4个转化子中已成功受到干扰。
2.4 转化子木质纤维素降解相关酶基因表达水平分析在得到的4个沉默子中挑选lelcrp1基因下调倍数最高的2个转化子lelcrp1-RNAi-11和lelcrp1-RNAi-13,分别提取总RNA,以反转录后cDNA为模板,采用荧光定量PCR技术检测部分木质纤维素降解相关酶基因在出发菌株W1以及RNAi转化子中的表达情况(表 2)。结果显示,在检测的26个木质纤维素降解相关酶基因中,有9个纤维素酶基因、1个半纤维素酶基因、2个辅助酶AA9基因和1个锰过氧化物酶基因的表达水平在这2个RNAi转化子中均呈现明显的下调趋势,另外13个酶基因表达则无明显变化。表明香菇异核菌株中lelcrp1基因的沉默影响了部分木质纤维素降解相关酶基因的表达水平,且对纤维素酶和锰过氧化物酶基因表达水平的影响较为显著。
| Gene ID | Gene family | Protein name | Relative quantification | ||
| W1(CK) | lelcrp1-RNAi-11 | lelcrp1-RNAi-13 | |||
| Cellulase | |||||
| LE01Gene04541 | GH12 | Endo-β-1, 4-glucanase | 1 | -66.1302 | -42.9299 |
| LE01Gene08136 | GH5 | Endo-β-1, 4-glucanase | 1 | -71.4921 | -13.9492 |
| LE01Gene11039 | GH5 | Glucan 1, 3-β-glucosidase | 1 | -1.6222 | -1.6050 |
| LE01Gene12502 | GH5 | Glucan 1, 3-β-glucosidase D | 1 | -1.2537 | -1.2874 |
| LE01Gene13053 | GH5 | Glucan 1, 3-β-glucosidase D | 1 | -1.2400 | -1.0985 |
| LE01Gene12864 | GH7 | Cellobiohydrolase Ⅰ | 1 | -3.3638 | -12.4348 |
| LE01Gene04089 | GH7 | Cellobiohydrolase Ⅰ | 1 | -8.4766 | -6.3056 |
| LE01Gene04829 | GH7 | Cellobiohydrolase Ⅰ | 1 | -62.1094 | -38.2943 |
| LE01Gene07961 | GH7 | Cellobiohydrolase Ⅰ | 1 | -38.0010 | -20.4303 |
| LE01Gene10050 | GH6 | Cellobiohydrolase Ⅱ | 1 | -33.7429 | -25.1632 |
| LE01Gene10512 | GH3 | β-glucosidase | 1 | -1.8679 | -6.1688 |
| LE01Gene10634 | GH3 | β-glucosidase | 1 | -34.7739 | -32.6943 |
| LE01Gene07574 | GH3 | β-glucosidase | 1 | -1.0480 | -2.0516 |
| LE01Gene10266 | AA9 | Lytic polysasccharide monooxygenase | 1 | -98.0890 | -39.8429 |
| LE01Gene09248 | AA9 | Lytic polysasccharide monooxygenase | 1 | -29.5640 | -45.0966 |
| Hemicellulase | |||||
| LE01Gene03223 | GH10 | Endo-1, 4-β-xylanase | 1 | -21.1862 | -17.7635 |
| LE01Gene07975 | GH10 | Endo-1, 4-β-xylanase | 1 | -1.1700 | -1.9892 |
| LE01Gene07762 | GH5 | Mannan endo-1, 4-β-mannosidase | 1 | 1.9330 | -1.0001 |
| LE01Gene00726 | GH5 | Mannan endo-1, 4-β-mannosidase | 1 | -1.2647 | 1.1469 |
| LE01Gene05156 | GH3 | Exo-1, 4-β-xylosidase | 1 | -1.1410 | -3.2034 |
| Lignin degrading protein | |||||
| LE01Gene09060 | AA2 | Manganese peroxidase | 1 | -24.0394 | -104.0387 |
| LE01Gene08997 | AA2 | Manganese peroxidase | 1 | -2.1837 | -1.9860 |
| LE01Gene04737 | AA1 | LACC 13 | 1 | -1.2489 | -1.6775 |
| LE01Gene04660 | AA1 | LACC 14 | 1 | -2.2591 | -2.3978 |
| LE01Gene04648 | AA1 | LACC 9 | 1 | -2.8776 | -2.7648 |
| LE01Gene02092 | AA5 | Glyoxal oxidase | 1 | -1.8333 | -2.9474 |
| *P < 0.01 and relative expression fold ≥4 represent significant difference. | |||||
2.5 转化子菌丝生长速度测定
在含有3.5 μg/mL潮霉素的MYG平板上,采用十字交叉法对4个沉默转化子的菌丝生长速度进行测量,用Microsoft Excel 2010软件进行数据处理和单因素方差分析(α=0.05),结果表明,转化子菌丝生长速度与出发香菇菌株W1相比具有显著性差异,转化子菌丝生长速度显著慢于W1 (表 3)。
| Strain | repetitions of hyphal average daily growth rate/mm | Average growth rate/mm | Error range/mm | P-value | ||||
| 1 | 2 | 3 | 4 | 5 | ||||
| W1 | 4.6375 | 4.6875 | 4.9375 | 4.8075 | 4.8725 | 4.7540 | ±0.1835 | 0.03 |
| lelcrp1-RNAi-11 | 4.4925 | 4.4525 | 4.6350 | 4.8100 | 4.7425 | 4.6265 | ±0.1835 | |
| lelcrp1-RNAi-12 | 4.4525 | 4.6000 | 4.5750 | 4.5950 | 4.7525 | 4.5950 | ±0.1575 | 0.02 |
| lelcrp1-RNAi-13 | 4.4800 | 4.4850 | 4.4800 | 4.1850 | 4.4400 | 4.4140 | ±0.2290 | 0.01 |
| lelcrp1-RNAi-14 | 4.3850 | 4.3375 | 4.5750 | 4.6200 | 4.2150 | 4.4265 | ±0.2115 | 0.01 |
| *P < 0.05 represent significant difference. | ||||||||
2.6 Southern杂交分析结果
以hpt557-F和hpt557-R为引物扩增hyg基因片段作为探针标记,以经限制性内切酶BamHⅠ酶切后的转化子基因组DNA为模板,Southern杂交结果显示,探针与出发菌株W1的酶切产物杂交无条带,而在受测的4个转化子中各出现一条带(图 4),表明外源插入性lelcrp1基因片段在香菇W1中以单拷贝的形式存在。

|
| 图 4 lelcrp1基因沉默转化子Southern杂交 Figure 4 Southern blotting of lelcrp1 silencing transformants. M: DNA-Hind Ⅲ; lane 1-5: W1, lelcrp1-RNAi-11, lelcrp1-RNAi-12, lelcrp1-RNAi-13, lelcrp1-RNAi-14. |
3 讨论
本研究采用根癌农杆菌介导转化法,获得了转录因子lelcrp1基因的4个沉默转化子,首次在香菇异核体菌株中对一个内源性HMG-box型转录因子lelcrp1实现了基因表达干扰。lelcrp1基因沉默子中多个木质纤维素酶基因表达水平出现了下降,而其所编码蛋白均为参与木质纤维素降解的重要蛋白[13],表明lelcrp1基因的表达变化影响了多个香菇木质纤维素降解相关酶基因的表达。LELCRP1蛋白中包含有DNA结合结构域,表明lelcrp1基因通过转录调控方式控制木质纤维素降解相关酶基因的表达。首次发现HMG-box结构域的转录因子能调控木质纤维素降解相关酶基因表达。
迄今为止,虽然子囊菌纤维素酶、半纤维素酶基因的表达调控得到了广泛研究,但白腐真菌调控木质纤维素降解相关酶基因的转录因子却鲜有研究报道。前人研究表明,不论是转录因子家族还是单个转录因子,在子囊菌和担子菌这两类真菌中都具有较大的差异,担子菌中仅少数转录因子与子囊菌具有同源性[22],担子菌相关转录因子研究仍然处于起步阶段。本文报道了在转录水平上调控多个木质纤维素相关酶基因表达的新型转录因子,这对香菇及其他担子菌相关转录因子的深入研究具有重要意义。
本研究通过农杆菌介导lelcrp1基因沉默片段转化香菇菌株W1后,插入片段以单拷贝形式存在,可以确定4个lehmo1基因沉默子中lelcrp1基因片段已经成功整合到了香菇基因组中。本研究同时也发现lelcrp1基因沉默子的菌丝生长速度比出发菌株慢,这一结果与Saloheimo等在红褐肉座菌(H. jecorina)敲除ace1基因导致菌丝在纤维素培养基上生长迟缓[23]的结果是类似的,因此推测lelcrp1基因沉默子菌丝生长速率降低是由于木质纤维素降解相关酶基因表达受到抑制造成的。
本研究通过RNAi技术初步研究了白腐真菌香菇转录因子基因lelcrp1的功能,发现此基因可影响香菇部分木质纤维素酶基因的表达水平,拓展了HMG-box型转录因子的功能,为进一步揭示香菇木质纤维素酶基因表达调控规律奠定了基础。
| [1] | Royse DJ, Sanchez JE. Ground wheat straw as a substitute for portions of oak wood chips used in shiitake (Lentinula edodes) substrate formulae. Bioresource Technology, 2007, 98(11): 2137-2141. DOI:10.1016/j.biortech.2006.08.023 |
| [2] | Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, Cate JHD, Glass NL. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(16): 6012-6017. DOI:10.1073/pnas.1118440109 |
| [3] | Nakazawa T, Izuno A, Kodera R, Miyazaki Y, Sakamoto M, Isagi Y, Honda Y. Identification of two mutations that cause defects in the ligninolytic system through an efficient forward genetics in the white-rot agaricomycete Pleurotus ostreatus. Environmental Microbiology, 2016, 19(1): 261-272. |
| [4] | Štros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cellular and Molecular Life Sciences, 2007, 64(19/20): 2590-2606. |
| [5] | Merz K, Hondele M, Goetze H, Gmelch K, Stoeckl U, Griesenbeck J. Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes & Development, 2008, 22(9): 1190-1204. |
| [6] | Ray S, Grove A. The yeast high mobility group protein HMO2, a subunit of the chromatin-remodeling complex INO80, binds DNA ends. Nucleic Acids Research, 2009, 37(19): 6389-6399. DOI:10.1093/nar/gkp695 |
| [7] | MacAlpine DM, Perlman PS, Butow RA. The high mobility group protein Abf2p influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proceedings of the National Academy of Sciences of the United States of America, 1998, 95(12): 6739-6743. DOI:10.1073/pnas.95.12.6739 |
| [8] | Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2010, 1799(1/2): 175-180. |
| [9] | Albert B, Colleran C, Léger-Silvestre I, Berger AB, Dez C, Normand C, Perez-Fernandez J, McStay B, Gadal O. Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-Box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Research, 2013, 41(22): 10135-10149. DOI:10.1093/nar/gkt770 |
| [10] | Dequard-Chablat M, Alland C. Two copies of mthmg1, encoding a novel mitochondrial HMG-like protein, delay accumulation of mitochondrial DNA deletions in Podospora anserine. Eukaryot Cell, 2002, 1(4): 503-513. DOI:10.1128/EC.1.4.503-513.2002 |
| [11] | Ohm RA, de Jong JF, de Bekker C, Wösten HA, Lugones LG. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Molecular Microbiology, 2011, 81(6): 1433-1445. DOI:10.1111/j.1365-2958.2011.07776.x |
| [12] |
Shi LL, van Peer AF, Guo L, Chen RL, Wang W, Yan JJ, Deng YJ, Xie BG. Agrobacterium-mediated transformation of an endogenous HMG-box transcription factor fvhom1 in Flammulina velutipes. Genomics and Applied Biology, 2014, 33(6): 1268-1274.
(in Chinese) 施乐乐, van PeerAF, 郭丽, 陈仁良, 王威, 严俊杰, 邓优锦, 谢宝贵. 农杆菌介导一个内源HMG-box转录因子fvhom1转化金针菇. 基因组学与应用生物学, 2014, 33(6): 1268-1274. |
| [13] | Cai YL, Gong YH, Liu W, Hu Y, Chen LF, Yan LL, Zhou Y, Bian YB. Comparative secretomic analysis of lignocellulose degradation by Lentinula edodes grown on microcrystalline cellulose, lignosulfonate and glucose. Journal of Proteomics, 2017, 163: 92-101. DOI:10.1016/j.jprot.2017.04.023 |
| [14] | Cho JH, Lee SE, Chang WB, Cha JS. Agrobacterium- mediated transformation of the winter mushroom, Flammulina velutipes. Mycobiology, 2006, 34(2): 104-107. DOI:10.4489/MYCO.2006.34.2.104 |
| [15] | Li C, Gong WB, Zhang L, Yang ZQ, Nong WY, Bian YB, Kwan HS, Cheung MK, Xiao Y. Association mapping reveals genetic loci associated with important agronomic traits in Lentinula edodes, shiitake mushroom. Frontiers in Microbiology, 2017, 8: 237. |
| [16] | Chen LF, Gong YH, Cai YL, Liu W, Zhou Y, Xiao Y, Xu ZY, Liu Y, Lei XY, Wang GZ, Guo MP, Ma XL, Bian YB. Genome sequence of the edible cultivated mushroom Lentinula edodes (Shiitake) reveals insights into lignocellulose degradation. PLoS One, 2016, 11(8): e0160336. DOI:10.1371/journal.pone.0160336 |
| [17] | Liu WZ, Xie YB, Ma JY, Luo XT, Nie P, Zuo ZX, Lahrmann U, Zhao Q, Zheng YY, Zhao Y, Xue Y, Ren J. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics, 2015, 31(20): 3359-3361. DOI:10.1093/bioinformatics/btv362 |
| [18] | Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genetics and Biology, 2004, 41(11): 973-981. DOI:10.1016/j.fgb.2004.08.001 |
| [19] | Dörnte B, Kües U. Fast microwave-based DNA extraction from vegetative mycelium and fruiting body tissues of agaricomycetes for PCR amplification. Current Trends in Biotechnology & Pharmacy, 2013, 7(4): 825-836. |
| [20] |
Liu XM, Wu XL, Chen Q, Qiu ZH, Zhang JX, Huang CY. Effects of heat stress on Pleurotus eryngii mycelial growth and its resistance to Trichoderma asperellum. Mycosystema, 2017, 36(11): 1566-1574.
(in Chinese) 刘秀明, 邬向丽, 陈强, 仇志恒, 张金霞, 黄晨阳. 高温胁迫对刺芹侧耳菌丝生长及其抗棘孢木霉能力的影响. 菌物学报, 2017, 36(11): 1566-1574. |
| [21] | Fan XZ, Zhou Y, Xiao Y, Bian YB. Cloning and characterization of two allelic glyceraldehyde-3-phosphate dehydrogenase genes in Auricularia auricula-judae. World Journal of Microbiology and Biotechnology, 2014, 30(1): 181-189. DOI:10.1007/s11274-013-1436-8 |
| [22] | Todd RB, Zhou M, Ohm RA, Leeggangers HA, Visser L, de Vries RP. Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genomics, 2014, 15: 214. DOI:10.1186/1471-2164-15-214 |
| [23] | Saloheimo A, Aro N, Ilmén M, Penttilä M. Isolation of the ace1 gene encoding a Cys2-His2 transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. The Journal of Biological Chemistry, 2000, 275(8): 5817-5825. DOI:10.1074/jbc.275.8.5817 |
 2018, Vol. 58
2018, Vol. 58