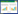中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 白晓晔, 钟智, 孙志宏, 张和平. 2018
- Xiaoye Bai, Zhi Zhong, Zhihong Sun, Heping Zhang. 2018
- 肠道中乳酸杆菌属特异性定量引物的筛选及验证
- Screening and evaluation of genus-specific primer for quantification of Lactobacillus in gut
- 微生物学报, 58(11): 1997-2010
- Acta Microbiologica Sinica, 58(11): 1997-2010
-
文章历史
- 收稿日期:2018-05-23
- 修回日期:2018-07-26
- 网络出版日期:2018-09-07
2. 内蒙古农业大学 农业农村部奶制品加工重点实验室, 内蒙古 呼和浩特 010018
2. Key Laboratory of Dairy Products Processing, Ministry of Agriculture, Inner Mongolia Agricultural University, Hohhot 010018, Inner Mongolia Autonomous Region, China
乳酸杆菌是兼性厌氧、无芽孢、生产乳酸的革兰氏阳性杆菌,它们是口腔、胃肠道和女性泌尿生殖道中的常见微生物[1]。乳酸杆菌通常被认为是非致病细菌,一些菌株还被用作益生菌以预防和治疗某些疾病[2-4]。乳酸杆菌与人和动物关系密切,常用它的含量变化作为健康与否的“风向标”。刘茜明等[5]应用q-PCR方法对葛根芩连汤治疗前后的巴马猪抗生素性腹泻动物模型肠道内乳酸杆菌数量的动态变化进行定量比较分析,发现葛根芩连汤治疗组的各肠段中乳酸杆菌属菌群含量呈显著上升趋势,对照组的含量显著下降,进而证明它具有调理肠道中乳酸杆菌的作用。张海波等[6]通过实时PCR技术对新生儿粪便中的双歧杆菌属、乳酸杆菌属进行定量检测,发现剖宫产组的粪便标本中乳酸杆菌数量低于自然分娩组。
定量乳酸杆菌属含量常用的方法是荧光实时定量PCR。然而,近几年,数字PCR凭借不依赖扩增曲线的Ct (循环阈值)、不受扩增效率影响、也不必采用内参基因和标准曲线等特点,成为了新一代绝对定量PCR的代表,其中尤以微滴式数字PCR应用较广[7-10]。但是,无论荧光实时定量PCR还是微滴式数字PCR,都离不开引物。理想的定量结果往往取决于引物的特异性,因此特异性较好的引物是定量成功的必要基础。理论上符合引物设计原则就能设计出理想的引物,实际上,不论是理论上绝对特异还是试验绝对特异,都很难设计出绝对完美的“引物”[11-12]。参考已有的引物,往往不能达到预期的特异性,而且引物质量参差不齐、特异性有待商榷,而设计引物必须经过大量试验进行特异性验证,费时费力,也不能保证引物的特异性。
本文通过查阅文献筛选出12对乳酸杆菌属或包含乳酸杆菌属的引物,根据文中提到的退火温度进行PCR,用琼脂糖凝胶电泳进行特异性检验,结果发现12对引物的特异性均不理想,因此,本文利用MEGA 6.0、Primer 5.0软件和NCBI网站BLAST软件对12对乳酸杆菌属引物的特异性和自身质量进行评估,以评估结果为依据进行理论筛选、组合和调整,获得理论特异性较好且符合乳酸杆菌属定量的引物组合,之后对新组合的引物进行特异性检验,筛选出特异性较好的乳酸杆菌属引物。最后借助微滴式数字PCR,用筛选好的引物对一份已知含有乳酸杆菌的粪便样品进行定量,通过先理论、后试验的方法,筛选出了一对乳酸杆菌属特异性引物,同时为快速筛选适宜的、特异性较好的引物提供理论依据。
1 材料和方法 1.1 主要菌株及DNA提取本试验用来检验引物特异性的16株细菌,包含14株模式菌株和2株非模式菌株,均由内蒙古农业大学乳品生物技术与工程教育部重点实验室提供,具体信息如表 1。利用天根生化科技(北京)有限公司生产的细菌基因组DNA提取试剂盒进行DNA提取,具体方法参照说明书进行。
| Type strain | Numbers |
| Enterococcus asini | DSM 11492 |
| Enterococcus faecium | ATCC 19434T |
| Enterococcus italicus | DSM 15952 |
| Lactobacillus casei | ATCC 334 |
| Lactobacillus salivarius | DSM 20555 |
| Lactobacillus helveticus | 1.2278 |
| Streptococcus thermophilus | NM-81-2 |
| Lactococcus plantarum | DSM 20686 |
| Weissella beninensis | DSM 22752 |
| Weissella confusa | IMAU 10245 |
| Weissella kandleri | DSM 20593 |
| Pediococcus acidilactici | JCM 8791 |
| Pediococcus pentosaceus | DSM 20336 |
| Leuconostoc psedomesenteroides | DSM 20193 |
| Bifidobacterium gallicum | DSM 20093 |
| Bifidobacterium longum subsp. infantis | DSM 20088 |
1.2 乳酸杆菌属16S rRNA基因序列的查找及引物的搜索
下载219种乳酸杆菌16S rRNA基因序列,汇成基因序列集。通过查阅文献搜索已有乳酸杆菌属和包含乳酸杆菌属的引物。将搜索到的引物借助MEGA 6.0以Lactobacillus salivarius的16S rRNA基因序列为模板(Accession number:AF089108.2)进行比对,确定每条引物的起始位置。
1.3 乳酸杆菌属引物的特异性试验将搜索到的12对乳酸杆菌属引物,以表 1中的16个亲缘相近种为模板,根据文中提到的最佳退火温度进行PCR扩增,利用琼脂糖凝胶检验其特异性。PCR反应体系、DNA Marker和核酸染料均购自大连宝生物技术有限公司;电泳仪(DYY-6D型)购自北京六一生物科技有限公司,凝胶成像仪(GDS-8000)购自美国UVP公司。
1.4 引物与乳酸杆菌属及其相近属的匹配率分析借助MEGA 6.0软件,将引物与乳酸杆菌属16S rRNA基因序列集及乳酸杆菌属相近的6个菌属(肠球菌属、乳球菌属、肠膜明串珠菌属、魏斯氏菌属、链球菌属和片球菌属)的16S rRNA基因序列进行比对,查看其与乳酸杆菌属及其相近属的匹配率(匹配率即与比对序列完全匹配的引物序列数/16S rRNA基因序列数)。比对时,正向引物的序列不变,反相引物的序列反相碱基互补。
1.5 乳酸杆菌属特异性引物筛选以表 2为依据,挑选:(1)只与乳酸杆菌属匹配的单条引物,(2)与其他菌属匹配但其匹配率远远低于乳酸杆菌属的引物,(3)与乳酸杆菌属和片球菌属匹配的引物,(4)与乳酸杆菌属、片球菌属和魏斯氏菌属匹配的引物。引物扩增子太短不仅很难与可能存在的二聚体区分,而且会影响引物扩增的特异性,而扩增子太长引物的扩增效率较短扩增子的低且扩增速率降低[13],因此本实验组合已经筛选出的引物单链,使引物的扩增产物片段大小介于100-300 bp。再借助MEGA 6.0比对引物3′端与乳酸杆菌属16S rRNA基因序列的匹配情况,挑选3′端与乳酸杆菌属16S rRNA序列完全匹配的引物。最后将符合上述所有条件的引物根据引物自身情况将序列做适当调整。
| Primer | Sequence (5′→3′) | Tm/℃ | Product size/bp | Environment |
| LactoF/LactoR | TGGAAACAGRTGCTAATACCG | 62 | 232 | Oral cavity[14-15],Cheese[16],Faeces[17] |
| GTCCATTGTGGAAGATTCCC | ||||
| Lac1/Lac2 | AGCAGTAGGGAATCTTCCA | 61 | 340 | Faeces[11, 18-19] |
| ATTYCACCGCTACACATG | ||||
| F-lacto/R-lacto | GAGGCAGCAGTAGGGAATCTTC | 60 | 126 | Faeces[20] |
| GGCCAGTTACTACCTCTATCCTTCTTC | ||||
| 27f/Lab-0677 | AGAGTTTGATCCTGGCTCAG | 56 | 700 | Faeces[21-22] |
| CACCGCTACACATGGAG | ||||
| Lab-0159f/Lab-0677r | GGAAACAG(A/G)TGCTAATACCG | 61 | 560 | Faeces[21],Breast milk[23-24] |
| CACCGCTACACATGGAG | ||||
| F/R | AGAGGAGAGTGGAACTCCATG | 55 | 205 | Vagina[25] |
| GTTAGCTGCAGCACTGAGAG | ||||
| LabF362/LabR677 | AGCAGTAGGGAATCTTCCA | 60 | - | Faeces[11],Milk products[26] |
| CACCGCTACACATGGAG | Faeces[21] | |||
| LACTO1-f/LACTO2 | GAGTTTGATCCTGGCTCAGGA | 56 | 200 | Strains[27] |
| CGGTATTAGCATCTGTTTCCA | ||||
| AM-LactoF/AM-LactoR | TGATGCATAGCCGAGTTGAG | 60 | - | Strains[28] |
| AGCCGAAACCCTTCTTCACT | ||||
| F-LBF7/R-LBR | ATGGAAGAACACCAGTGGCG | 50 | - | Vagina[29-30] |
| CAGCACTGA GAG GCG GAA AC | ||||
| WLAB1/WLAB2 | TCCGGATTTATTGGGCGTAAAGCGA | 60 | 400 | Red wine[12] |
| TCGAATTAAACCACATGCTCCA | ||||
| Lacb-0018/Lacb-336 | TGCCTAATACATGCAAGTCGA | 52 | 318 | Faeces[31] |
| GTTTGGGCCGTGTCTCAGT | ||||
| -: no mention in the text. | ||||
1.6 引物的特异性验证
将经筛选、组合和调整的引物,以乳酸杆菌亲缘相近的16株菌为模板,退火温度60 ℃,采用1.0%琼脂糖电泳对PCR扩增产物特异性进行检测,并用凝胶成像仪观察检测结果。
1.7 微滴式数字PCR反应将2 μL DNA、上下游引物各0.2 μL、10.0 μL 2×ddPCR SuperMix和7.6 μL的灭菌超纯水混合,制备成ddPCR反应液。将ddPCR反应液与微滴生成油分别加至微滴发生卡的对应孔,放入微滴生成仪(反应所用试剂均购自美国Bio-Rad公司)。使用EvaGreen程序进行PCR扩增,退火温度60 ℃。将PCR扩增后的96孔板置于微滴分析仪进行检测和分析。
1.8 乳酸杆菌属引物在数字PCR定量中的应用借助QX200 Droplet Digital PCR系统(美国Bio-Rad公司),使用EvaGreen进行PCR扩增,退火温度60 ℃,对引物Lab-F1/Lab-R1依次进行如下试验:(1)依次以L. salivarius、W. beninensis、P. acidilactici、L. psedomesenteroides、S. thermophilus、L. plantarum和灭菌超纯水为扩增模板,检验引物Lab-F1/Lab-R1的特异性;(2)将L. salivarius的DNA稀释液按10倍梯度依次稀释,检验Lab-F1/ Lab-R1的灵敏性;(3)以Lab-F1/Lab-R1为引物,对不同的粪便中的乳酸杆菌进行检测。
2 结果和分析 2.1 现有乳酸杆菌引物信息通过查阅文献共找到12对基于乳酸杆菌属16S rRNA基因序列保守区设计的引物,详细信息见表 2。
将搜索到的引物借助MEGA 6.0以L. salivarius的16S rRNA基因序列为模板进行比对,每条引物的起始位置如图 1。其中,黑色方框从左到右表示16S rRNA基因序列的可变区V1-V9;图中箭头表示引物的位置,同一行为一对引物,其中每行中靠左位置的箭头代表正向引物的起始位置,靠右位置的箭头代表反向引物的起始位置。
|
| 图 1 引物在Lactobacillus salivarius 16S rRNA基因序列(Accession number: AF089108.2)上的位置 Figure 1 The relative position of the primers on the L. salivarius 16S rRNA gene sequence (Accession number: AF089108.2). |
通过图 1发现,12对引物的起始位置多数集中在V1-V6区间,其中27f、LACTO1-f和Lacb-0018这3条引物位于V1区之前的保守区,LactoF、LactoR、Lac1、F-lacto、R-lacto、Lab-0159f、LabF362、LACTO2、Lacb-336、AM-LactoF和AM-LactoR集中在V2-V3区,Lac2、Lab-0677r (Lab-0677)、F、R、LabR677、F-LBF7和R-LBR集中V4-V5区,WLAB1位于V4区之前的保守区,WLAB2位于V6区前的保守区。
2.2 现有乳酸杆菌属引物特异性试验结果以乳酸杆菌属及其亲缘关系相近的7个属共16株细菌DNA为模板,通过琼脂糖凝胶电泳对12对引物的特异性进行检验,检验结果如图 2。其中,A-L依次代表F/R、27f/Lab-0677、LACTO1-f/ LACTO2r、F-lacto/R-lacto、Lab-0159f/Lab-0677r、LactoF/LactoR、F-LBF/R-LBR、Lacb-0018/Lacb-336、Lac1/Lac2、LabF362/LabR67、AM-LactoF/AM-LactoR和WLAB1/WLAB2搜索到的12对引物。每对引物对应的模板一致,从左到右依次为E. asini、E. italicus、E. faecium、W. beninensis、W. cnofusa、W. kandleri、P. acidilactici、P. pentosaceus、L. salivarius、L. helveticus、L. casei、L. psedomesenteroides、L. plantarum、S. thermophilus、B. gallicum和B. longum subsp. infantis。

|
| 图 2 引物特异性验证结果 Figure 2 Specificity of searched primers. A-L denote F/R, 27f/Lab-0677, LACTO1-f/LACTO2r, F-lacto/R-lacto, Lab-0159f/Lab-0677r, LactoF/LactoR, F-LBF/R-LBR, Lacb-0018/Lacb-336, Lac1/Lac2, LabF362/LabR67, AM-LactoF/AM-LactoR and WLAB1/WLAB2. 1-16 denote E. asini, E. faecium, E. italicus, L. casei, L. salivarius, L. helveticus, W. beninensis, W. cofusa, W. kandleri, P. acidilactici, P. pentosaceus, L. psedomesenteroides, S. thermophiles, L. plantarum, B. gallicum and B. longum subsp. infantis, respectively. |
在本试验环境下,27f/Lab-0677、LACTO1-f/ LACTO2r、Lab-0159f/Lab-0677r、F-LBF/R-LBR、WLAB1/WLAB2这5对引物可以扩增出包含乳酸杆菌在内的全部菌株。F/R、LactoF/LactoR、Lacb-0018/Lacb-336、LabF362/LabR67和AM-LactoF/ AM-LactoR这5对引物可以扩增出包含乳酸杆菌在内的大部分模板,而F-lacto/R-lacto和Lac1/Lac2无法扩增出乳酸杆菌。每对引物或者特异性不太理想或者扩增效果不太理想,均难以符合乳酸杆菌定量的要求。
2.3 引物与乳酸杆菌属及其相近属的匹配率12对引物均与乳酸杆菌属的匹配程度不同,其中9条引物与乳酸杆菌属的匹配率达90%以上。12对引物中大部分引物还与片球菌属和魏斯氏菌属有较高的匹配率。12对引物中,部分引物与其他4个属的部分细菌种匹配,具体匹配率详见表 3。
| Primers | Lactobacillus | Streptococcus | Leuconostoc | Pediococcus | Lactococcus | Weissella | Enterococcus |
| LactoF | 86.80 | - | 100.00 | 80.00 | - | 45.50 | 100.00 |
| LactoR | 81.70 | - | - | 86.70 | - | 50.00 | - |
| F-lacto | 99.50 | 100.00 | - | 86.70 | 100.00 | 100.00 | 100.00 |
| R-lacto: | 7.80 | - | - | - | - | - | - |
| Lab-0159f | 87.20 | - | 100.00 | 86.70 | - | 54.50 | 100.00 |
| Lab-0677r | 96.80 | - | 100.00 | 86.70 | - | 95.50 | - |
| 27f | 20.10 | 24.80 | 10.50 | - | 20.00 | - | 16.70 |
| Lab-0677r | 96.80 | - | 100.00 | 86.70 | - | 95.50 | - |
| F | 37.40 | - | - | 30.70 | - | 31.80 | - |
| R | 14.20 | - | - | - | - | - | - |
| LACTO1-f | 22.00 | 24.80 | 10.50 | - | 20.00 | 4.54 | 16.70 |
| LACTO2-r | 37.40 | - | 100.00 | 92.30 | - | 45.50 | - |
| F-LBF | 92.70 | - | 57.90 | 84.60 | - | 63.60 | - |
| R- LBR | 13.20 | - | - | - | - | - | - |
| WLAB1 | 74.90 | 93.10 | 100.00 | 86.70 | 100.00 | 95.50 | 100.00 |
| WLAB2 | 100.00 | 100.00 | 100.00 | 86.70 | 86.70 | 100.00 | 100.00 |
| lac1 | 99.50 | - | - | 86.70 | - | 100.00 | - |
| lac2 | 99.50 | 100.00 | 100.00 | 86.70 | 100.00 | 100.00 | 100.00 |
| LabF362 | 99.50 | - | - | 86.70 | - | 100.00 | - |
| LabR677 | 96.80 | - | 100.00 | 86.70 | - | 95.50 | - |
| AM-LactoF | 15.50 | - | 100.00 | - | - | 50.00 | - |
| AM-LactoR | 32.40 | - | - | 46.20 | - | - | - |
| Lacb-0018 | 75.80 | - | 84.20 | 46.20 | 20.00 | 81.80 | 70.40 |
| Lacb-336 | 76.30 | 1.98 | 100.00 | 7.69 | 86.70 | - | 35.20 |
| -: no match. | |||||||
2.4 乳酸杆菌属特异性引物的筛选结果
以表 3为依据,共筛选出符合条件的16条引物:R-lacto,AM-LactoR,LactoR,LabF362,AM-LactoF,Lab-0677r,LACTO1f,R-LBR,F-LBF,F,R,lac1和lac2。参考图 1组合扩增片段在100-300 bp的引物对11对。最后根据引物3′端与乳酸杆菌属16S rRNA基因序列的匹配情况,从这11对组合引物中筛选出了3对3′端与乳酸杆菌属完全匹配的引物:LactoR/F,LabF362/F和AM-LactoF/ R-lacto。将这3对引物调整正反向,得到新的引物对序列,重新依次命名为Lab-F1/Lab-R1、Lab-F2/Lab-R2、Lab-F3/Lab-R3,其序列如下:Lab-F1/Lab-R1 (5′-GGGAATCTTCCACAATGGAC- 3′/5′-CATGGAGTTCCACTCTCCTCT-3′),Lab-F2/Lab- R2 (5′-AGCAGTAGGGAATCTTCCA-3′/5′-CATGGA GTTCCACTCTCCTCT-3′),Lab-F3/Lab-R3 (5′-TGAT GCATAGCCGAGTTGAG-3′/5′-GGCCAGTTACTACCTCTATCCTTCTTC-3′)。
| Type strains | Primers | ||||||
| Lab-F1/ Lab-R1 | Lab-F2/ Lab-R2 | Lab-F3/ Lab-R3 | |||||
| E. asini | - | - | - | ||||
| E. faecium | - | - | - | ||||
| E. italicus | - | - | - | ||||
| L. casei | + | + | - | ||||
| L. salivarius | + | + | - | ||||
| L. helveticus | + | + | + | ||||
| W. beninensis | + | + | - | ||||
| W. confusa | + | + | + | ||||
| W. kandleri | + | + | - | ||||
| P. acidilactici | + | + | - | ||||
| P. pentosaceus | + | - | - | ||||
| L. psedomesenteroides | - | + | - | ||||
| S. thermophilus | - | + | - | ||||
| L. plantarum | - | - | - | ||||
| B. gallicum | - | - | - | ||||
| B. longum subsp. infantis | - | - | - | ||||
| +: amplification; -: no amplification. | |||||||

|
| 图 3 组合乳酸杆菌属引物特异性验证结果 Figure 3 Specificity of new Lactobacillus genus-specific primers evaluated by agarose gel electrophoresis. 1-16 denote E. asini, E. faecium, E. italicus, L. casei, L. salivarius, L. helveticus, W. beninensis, W. confusa, W. kandleri, P. acidilactici, P. pentosaceus, L. psedomesenteroides, S. thermophilus, L. plantarum, B. gallicum and B. longum subsp. infantis, respectively. |
2.5 引物对的特异性验证结果
用理论筛选组合好的3对乳酸杆菌属引物扩增表 1中的16株细菌DNA,将扩增产物进行琼脂糖凝胶电泳,试验结果如表 4和图 3。
通过表 4和图 3可知,引物Lab-F3/Lab-R3仅扩增L. helveticus和W. cofusa呈阳性;Lab-F2/ Lab-R2除了扩增所有预期菌种L. casei、L. salivarius、L. helveticus、W. beninensis、W. confusa、W. kandleri、P. acidilactici、P. pentosaceus呈阳性外,扩增L. psedomesenteroides和S. thermophilus也呈阳性;而Lab-F1/Lab-R1仅扩增所有预期菌种L. casei、L. salivarius、L. helveticus、W. beninensis、W. confusa、W. kandleri、P. acidilactici和P. pentosaceus呈阳性。因此,乳酸杆菌属引物Lab-F1/Lab-R1的扩增效果和特异性较好,将这对引物用Lab1表示。
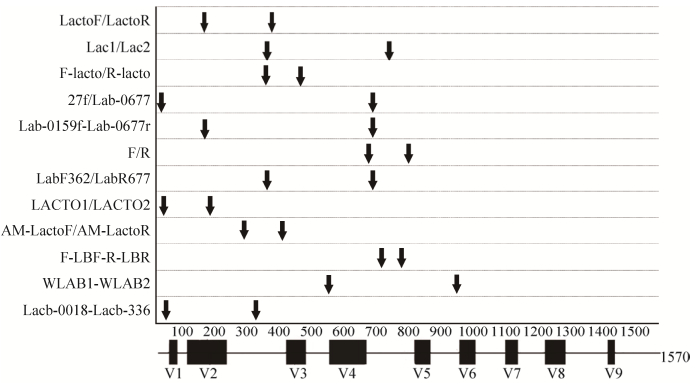
2.6 乳酸杆菌引物在数字PCR定量中的应用 2.6.1 乳酸杆菌属引物特异性检验结果: 以1株乳酸杆菌模式菌、6株乳酸杆菌亲缘模式菌和1份灭菌超纯水为扩增模板,借助QX200 Droplet Digital PCR系统检验引物Lab1的特异性。结果发现,仅L. salivarius出现大量阳性微滴,且阳性微滴和阴性微滴明显分开,见图 4。
|
| 图 4 ddPCR特异性检验结果 Figure 4 Specificity of primer Lab1. One positive and seven negative templates were detected using ddPCR. A04-H04 denote the type bacterial DNA of L. salivarius, E. asini, S. thermophiles, L. plantarum, W. beninensis, P. acidilactici and L. psedomesenteroides, respectively. |
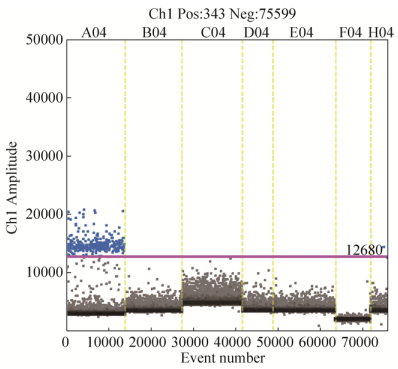
2.6.2 乳酸杆菌属引物灵敏度检验: 以按10倍梯度依次稀释的L. salivarius DNA稀释液为模板,借助QX200 Droplet Digital PCR系统检验Lab1的灵敏性,结果发现不同浓度的L. salivarius拷贝数依次为3780.0、381.0、40.2、7.5 copies/μL,阳性微滴数随着浓度降低呈线性递减趋势,见图 5。

|
| 图 5 ddPCR灵敏性检验结果 Figure 5 Sensitivity of Lab-1. DNA diluents of L. salivarius with different concentrations were detected using ddPCR. A03-D03 denote 134.7000, 13.4700, 1.3470, 0.1347 ng/μL of L. salivarius DNA, respectively. |
2.6.3 乳酸杆菌属引物实用性检验结果: 借助QX200 Droplet Digital PCR系统对3份健康成人粪便和3份婴儿胎便的DNA稀释液进行检测。以Lab1为引物,使用EvaGreen进行PCR扩增,退火温度60 ℃。ddPCR结果显示,6份粪便样品(A-F)中的双歧杆菌拷贝数依次为4.2、102.0、9.5、7.9、3.9、67.4 copies/μL,同时Lab1这对引物可以明显区分阴性微滴与阳性微滴,即其特异性较好,见图 6。
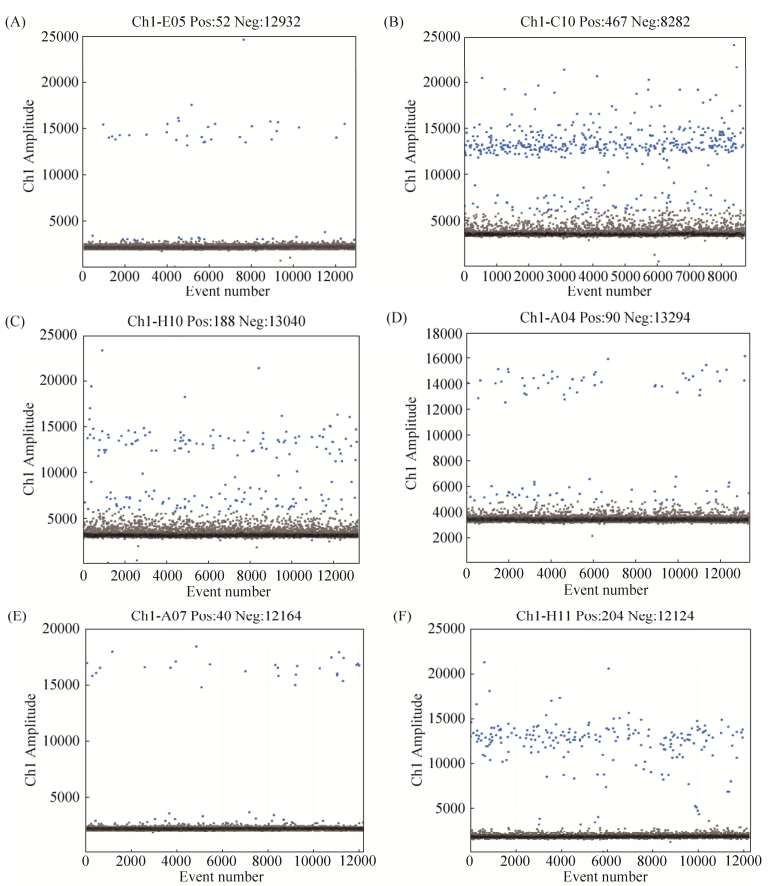
|
| 图 6 ddPCR有效性检测结果 Figure 6 Efficiency of Lab1. Six different fecal DNA dilutions were used to test the efficiency of Lab1 using ddPCR. A-F denote six different fecal samples respectively. |
3 分析和展望
人体乳酸杆菌的存在及其数量,往往被看作身体健康的“晴雨表”[32-34]。在定量过程中引物不可缺少,本实验经过理论分析初筛和琼脂糖凝胶试验筛选出1对特异性较好的乳酸杆菌属特异性引物,命名为Lab-F1/Lab-R1 (Lab1)。随后将Lab1作为EvaGreen ddPCR扩增的引物,借助QX200 Droplet Digital PCR系统依次对引物进行了特异性、灵敏性和有效性检验,结果发现这对引物不仅具有较好的特异性(图 4)和灵敏性(图 5),还可以用来有效定量粪便中的乳酸杆菌(图 6)。
乳酸杆菌引物设计是基于其特有的序列设计引物,这种所谓的特有序列是针对目前已知的序列,而未知的序列难以预测,这就导致实际应用中难以保证其特异性,尤其是复杂样品中乳酸杆菌的检测。Byun等[14]以35种乳酸杆菌的16S rRNA基因序列为参考设计的乳酸杆菌属引物,可以对龋齿中存在的乳酸杆菌进行定量分析,Delroisse等[20]设计的乳酸杆菌属引物可以扩增大鼠粪便中的15种乳酸杆菌,Heilig等[21]分析肠道微生物多样性所用的乳酸杆菌属引物一条是基于乳酸杆菌属设计,另一条引物却可以扩增出乳酸杆菌属和肠球菌属,还有Isabel等[12]设计的乳酸菌引物可以同时扩增出乳酸杆菌属、链球菌属、片球菌属和乳球菌属,而Walter等[11]设计的引物可以同时扩增出乳酸杆菌属、片球菌属、明串球菌属和魏斯氏菌属。这些引物往往都有其最佳使用环境,或者扩大了检测范围,然而,目前很难设计一对引物可以特异性地扩增出所有乳酸杆菌。无论是重新设计乳酸杆菌属引物,还是参考文献中提供的引物,都存在不可避免的缺陷,而本文成功地通过先理论筛选后试验验证的方法筛选出适合检测粪便中乳酸杆菌的特异性乳酸杆菌属引物,进而证明这种理论与实验结合的引物筛选的方法是可行的。
由于乳酸杆菌是非单系起源、且与其他乳酸菌基因序列具有高度相似性[35],本试验以16株标准菌株为模板进行引物特异性验证时,Lab1这对引物不仅扩增出了所有乳酸杆菌,还扩增出了魏斯氏菌和片球菌。根据目前粪便微生物多样性分析发现,粪便中的片球菌和魏斯氏菌属相对含量比较低,基本检测不到,往往不做微观研究[36-39],这就减少了定量过程中对乳酸杆菌属真实含量的影响。同时,通过NCBI网站(https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/Pediococcus;https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/lactobacillus)查询片球菌和乳酸杆菌属全基因组信息可以发现,片球菌的G+C含量普遍较乳酸杆菌属的低,定量乳酸杆菌属时,可适当将退火温度提高,以提高引物的特异性。此外寻找乳酸杆菌属特有的基因序列,如核心基因等,作为靶序列设计属水平特异性引物也许是引物设计的一条新途径。
| [1] | Sherid M, Samo S, Sulaiman S, Husein H, Sifuentes H, Sridhar S. Liver abscess and bacteremia caused by lactobacillus: role of probiotics? Case report and review of the literature. BMC Gastroenterology, 2016, 16: 138. DOI:10.1186/s12876-016-0552-y |
| [2] | Chaves BD, Brashears MM, Nightingale KK. Applications and safety considerations of Lactobacillus salivarius as a probiotic in animal and human health. Journal of Applied Microbiology, 2017, 123(1): 18-28. DOI:10.1111/jam.2017.123.issue-1 |
| [3] | Santarmaki V, Kourkoutas Y, Zoumpopoulou G, Mavrogonatou E, Kiourtzidis M, Chorianopoulos N, Tassou C, Tsakalidou E, Simopoulos C, Ypsilantis P. Survival, intestinal mucosa adhesion, and immunomodulatory potential of Lactobacillus plantarum strains. Current Microbiology, 2017, 74(9): 1061-1067. DOI:10.1007/s00284-017-1285-z |
| [4] |
Liu XM, Peng ZR, Ni XQ, Yang J, Zou ZY, Qiu CH, Zeng D, Yang LG. Aperient effect of Fructooligosaccharides and Lactobacillus on constipation model of rats. Food Science, 2013, 34(11): 296-299.
(in Chinese) 刘晓梅, 彭芝榕, 倪学勤, 杨杰, 邹昭勇, 邱春红, 曾东, 杨立格. 低聚果糖、乳酸杆菌对便秘模型大鼠的通便功能影响. 食品科学, 2013, 34(11): 296-299. DOI:10.7506/spkx1002-6630-201311063 |
| [5] |
Liu QM, Yang GY, He GZ, Tian WY, Cai K, Wang P, Huang G, Liang JD, Wang WJ. Influence of gegenqinlian decoction on intestinal Lactobacillus in antibiotic associated diarrhea model treatment. Acta Ecologae Animalis Domastici, 2016, 37(11): 74-78.
(in Chinese) 刘茜明, 杨光勇, 何光志, 田维毅, 蔡琨, 王平, 黄高, 梁建东, 王文佳. 葛根芩连汤对抗生素相关腹泻肠道乳酸杆菌属的影响研究. 家畜生态学报, 2016, 37(11): 74-78. DOI:10.3969/j.issn.1673-1182.2016.11.013 |
| [6] |
Zhang HB, Yu JL, Ai Q, Xu YZ. Impact of cesarean section delivery on intestinal Bifidobacteria and lactobacillius in newborn babies in the first three days after delivery. Chongqing Medicine, 2012, 41(21): 2157-2159.
(in Chinese) 张海波, 余加林, 艾青, 徐艳珍. 剖宫产对新生儿出生后3d内肠道双歧杆菌和乳酸杆菌定植的影响. 重庆医学, 2012, 41(21): 2157-2159. DOI:10.3969/j.issn.1671-8348.2012.21.014 |
| [7] | Zhao Y, Xia QY, Yin YP, Wang ZK. Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri Subsp. citri. PLoS One, 2016, 11(7): e0159004. DOI:10.1371/journal.pone.0159004 |
| [8] | Iwobi A, Gerdes L, Busch U, Pecoraro S. Droplet digital PCR for routine analysis of genetically modified foods (GMO) - a comparison with real-time quantitative PCR. Food Control, 2016, 69: 205-213. DOI:10.1016/j.foodcont.2016.04.048 |
| [9] | Verhaegen B, de Reu K, De Zutter L, Verstraete K, Heyndrickx M, van Coillie E. Comparison of droplet digital PCR and qPCR for the quantification of Shiga toxin-producing Escherichia coli in bovine feces. Toxins (Basel), 2016, 8(5): 157. DOI:10.3390/toxins8050157 |
| [10] | Cao YP, Raith MR, Griffith JF. Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Research, 2015, 70: 337-349. DOI:10.1016/j.watres.2014.12.008 |
| [11] | Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Applied and Environmental Microbiology, 2001, 67(6): 2578-2585. DOI:10.1128/AEM.67.6.2578-2585.2001 |
| [12] | Lopez I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, Marshall M, VanderGheynst J, Mills DA. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Applied and Environmental Microbiology, 2003, 69(11): 6801-6807. DOI:10.1128/AEM.69.11.6801-6807.2003 |
| [13] | 林万明. PCR技术操作和应用指南. 北京: 人民军医出版社, 1993: 15. |
| [14] | Byun R, Nadkarni MA, Chhour KL, Martin FE, Jacques NA, Hunter N. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. Journal of Clinical Microbiology, 2004, 42(7): 3128-3136. DOI:10.1128/JCM.42.7.3128-3136.2004 |
| [15] | Vacharaksa A, Suvansopee P, Opaswanich N, Sukarawan W. PCR detection of Scardovia wiggsiae in combination with Streptococcus mutans for early childhood caries-risk prediction. European Journal of Oral Sciences, 2015, 123(5): 312-318. DOI:10.1111/eos.12208 |
| [16] | Fonseca S, Ouoba LⅡ, Franco I, Carballo J. Use of molecular methods to characterize the bacterial community and to monitor different native starter cultures throughout the ripening of Galician chorizo. Food Microbiology, 2013, 34(1): 215-226. DOI:10.1016/j.fm.2012.12.006 |
| [17] | McOrist AL, Jackson M, Bird AR. A comparison of five methods for extraction of bacterial DNA from human faecal samples. Journal of Microbiological Methods, 2002, 50(2): 131-139. DOI:10.1016/S0167-7012(02)00018-0 |
| [18] | Di Cagno R, Rizzello CG, Gagliardi F, Ricciuti P, Ndagijimana M, Francavilla R, Guerzoni ME, Crecchio C, Gobbetti M, de Angelis M. Different fecal microbiotas and volatile organic compounds in treated and untreated children with celiac disease. Applied and Environmental Microbiology, 2009, 75(12): 3963-3971. DOI:10.1128/AEM.02793-08 |
| [19] | Ritchie LE, Burke KF, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS. Characterization of fecal microbiota in cats using universal 16S rRNA gene and group-specific primers for Lactobacillus and Bifidobacterium spp. Veterinary Microbiology, 2010, 144(1/2): 140-146. |
| [20] | Delroisse JM, Boulvin AL, Parmentier I, Dauphin RD, Vandenbol M, Portetelle D. Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiological Research, 2008, 163(6): 663-670. DOI:10.1016/j.micres.2006.09.004 |
| [21] | Heilig HGHJ, Zoetendal EG, Vaughan EE, Marteau P, Akkermans ADL, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Applied and Environmental Microbiology, 2002, 68(1): 114-123. DOI:10.1128/AEM.68.1.114-123.2002 |
| [22] | Nielsen DS, Møller PL, Rosenfeldt V, Paerregaard A, Michaelsen KF, Jakobsen M. Case study of the distribution of mucosa-associated Bifidobacterium species, Lactobacillus species, and other lactic acid bacteria in the human colon. Applied and Environmental Microbiology, 2003, 69(12): 7545-7548. DOI:10.1128/AEM.69.12.7545-7548.2003 |
| [23] | Collado MC, Delgado S, Maldonado A, Rodríguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Letters in Applied Microbiology, 2009, 48(5): 523-528. DOI:10.1111/lam.2009.48.issue-5 |
| [24] | Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L. Lactobacilli and bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. Journal of Pediatric Gastroenterology and Nutrition, 2014, 59(1): 78-88. DOI:10.1097/MPG.0000000000000347 |
| [25] | Doi M, Gamo S, Okiura T, Nishimukai H, Asano M. A simple identification method for vaginal secretions using relative quantification of Lactobacillus DNA. Forensic Science International: Genetics, 2014, 12: 93-99. DOI:10.1016/j.fsigen.2014.05.005 |
| [26] | Kim DH, Chon JW, Kim H, Kim HS, Choi D, Hwang DG, Seo KH. Detection and enumeration of lactic acid bacteria, acetic acid bacteria and yeast in kefir grain and milk using quantitative real-time PCR. Journal of Food Safety, 2015, 35(1): 102-107. DOI:10.1111/jfs.2015.35.issue-1 |
| [27] | Chagnaud P, Machinis K, Coutte LA, Marecat A, Mercenier A. Rapid PCR-based procedure to identify lactic acid bacteria: application to six common Lactobacillus species. Journal of Microbiological Methods, 2001, 44(2): 139-148. DOI:10.1016/S0167-7012(00)00244-X |
| [28] | Henriques A, Cereija T, Machado A, Cerca N. In silico vs in vitro analysis of primer specificity for the detection of Gardnerella vaginalis, Atopobium vaginae and Lactobacillus spp. BMC Research Notes, 2012, 5: 637. DOI:10.1186/1756-0500-5-637 |
| [29] | Zariffard MR, Saifuddin M, Sha BE, Spear GT. Detection of bacterial vaginosis-related organisms by real-time PCR for Lactobacilli, Gardnerella vaginalis and Mycoplasma hominis. FEMS Immunology & Medical Microbiology, 2002, 34(4): 277-281. |
| [30] | Jespers V, Menten J, Smet H, Poradosú S, Abdellati S, Verhelst R, Hardy L, Buvé A, Crucitti T. Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiology, 2012, 12: 83. DOI:10.1186/1471-2180-12-83 |
| [31] | Rekha R, Rizvi MA, Jaishree P. Designing and validation of genus-specific primers for human gut flora study. Electronic Journal of Biotechnology, 2006, 9(5): 505-511. |
| [32] | Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Applied and Environmental Microbiology, 2006, 72(4): 2359-2365. DOI:10.1128/AEM.72.4.2359-2365.2006 |
| [33] | Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, Dong M, Chen YZ, Ismail N, Ndung'u T, Ghebremichael MS, Wesemann DR, Mitchell C, Dong KL, Huttenhower C, Walker BD, Virgin HW, Kwon1 DS. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young south african women. Immunity, 2017, 46(1): 29-37. |
| [34] | Xi XX, Huang WQ, Hou QC, Kwok LY, Sun ZH, Sun TS. Assessment of microbial community structure in human colostrum and mature milk based on geographical location and delivery mode. Science Bulletin, 2017, 62(11): 745-747. DOI:10.1016/j.scib.2017.03.016 |
| [35] | Simpson JM, McCracken VJ, Gaskins HR, Mackie RI. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Applied and Environmental Microbiology, 2000, 66(11): 4705-4714. DOI:10.1128/AEM.66.11.4705-4714.2000 |
| [36] | Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, Marmon S, Neimann A, Brusca S, Patel T, Manasson J, Pamer EG, Littman DR, Abramson SB. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis & Rheumatology, 2015, 67(1): 128-139. |
| [37] | Opstelten JL, Plassais J, Van Mil SWC, Achouri EM, Pichaud M, Siersema PD, Oldenburg B, Cervino ACL. Gut microbial diversity is reduced in smokers with crohn's disease. Inflammatory Bowel Diseases, 2016, 22(9): 2070-2077. DOI:10.1097/MIB.0000000000000875 |
| [38] | Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in crohn's disease revealed by a metagenomic approach. Gut, 2006, 55(2): 205-211. DOI:10.1136/gut.2005.073817 |
| [39] | Machiels K, Sabino J, Vandermosten L, Joossens M, Arijs I, de Bruyn M, Eeckhaut V, van Assche G, Ferrante M, Verhaegen J, van Steen K, van Immerseel F, Huys G, Verbeke K, Wolthuis A, de Buck van Overstraeten A, D'Hoore A, Rutgeerts P, Vermeire S. Specific members of the predominant gut microbiota predict pouchitis following colectomy and IPAA in UC. Gut, 2017, 66(1): 79-88. DOI:10.1136/gutjnl-2015-309398 |
 2018, Vol. 58
2018, Vol. 58