中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 王鹏, 吴群, 徐岩. 2018
- Peng Wang, Qun Wu, Yan Xu. 2018
- 中国白酒发酵过程中的核心微生物群及其与环境因子的关系
- Core microbiota in Chinese liquor fermentation and associations with environmental factors
- 微生物学报, 58(1): 142-153
- Acta Microbiologica Sinica, 58(1): 142-153
-
文章历史
- 收稿日期:2017-02-14
- 修回日期:2017-03-17
- 网络出版日期:2017-04-06
多菌种参与的固态发酵被认为是一种古老、经济且对人体有益的食品发酵技术[1]。由于多种微生物共同作用并具有复杂的代谢活力,这种类型的发酵可以产生独特的食品风味和口感特征[2]。通过寻找核心微生物群有利于人工构建合成微生物群落(synthetic microbial communities)并获得易操作且可重复的发酵系统[3-4]。目前从系统水平上寻找核心微生物并构建模拟的微生物群落来研究群落的性能和稳定性是常用的方法[5]。有研究者发现,不同国家的奶酪中具有稳定且丰度较高的微生物属,如Staphylococcus、Brevibacterium、Brachybacterium、Penicillium和Candida等。通过筛选相应的物种并人工构建奶酪皮,证明了奶酪发酵过程中微生物群落聚集的动态变化具有一定的可操控性和可重复性[6]。
寻找复杂发酵系统中的核心微生物群是构建模拟微生物群落的关键问题之一。目前寻找核心微生物群的方法可以归纳为以下几种:(1)功能相关的微生物(functional microbiota)[7-8],如通过微生物群落和代谢物的相关性分析在食醋发酵中找到了由7个属组成的功能微生物群[8];(2)丰度界定的优势微生物(dominant microbiota)[6, 9],如Hu把平均相对丰度>1%且至少出现在一个样品以上的属定义为窖泥中的核心微生物属,共鉴定出17个核心属[9];(3)在样本中分布频率较大的普遍微生物群(ubiquitous microbiota)[10-11],如Rui等将至少分布在90%的家用沼气池中的OTU定义为核心OTU[10];(4)共现性网络分析出共现的微生物群(co-occurring microbiota)[12-13],如Barberan等根据微生物的同现率(incidence of co-occurrence)找出了土壤群落中的2个关键门为Verrucomicrobia和Acidobacteria[14]。
本研究以中国传统的白酒发酵为研究载体,试图尝试多种方法以更全面地揭示该发酵系统的核心微生物群(core microbiota)并探究环境因素对其的影响。通过高通量扩增子测序深入挖掘发酵过程中的微生物群落结构,GC-MS测定样品中的挥发性化合物含量,并使用多元统计学分析解析发酵过程中微生物群落和代谢轮廓的变化规律,找出了白酒发酵过程中的核心微生物群,并量化了环境因素对核心微生物群的影响,为白酒生产的定向调控提供一定的理论基础。
1 材料和方法 1.1 样品的采集样品从江苏北部某酒厂采集,分别从2个窖池发酵的0、5、8、10、15、20、30、40 d取样,共计16份。取样时间为2015年4月。样品采集,一式2份,立即密封。一份–20 ℃保存用来测定理化指标和物质含量;一份–80 ℃保存用来提取DNA。
1.2 理化指标、乙醇、乳酸和挥发性化合物的测定使用温度计插入窖池1 m深处测定酒醅温度。酒醅中的水分采用恒温干燥法测定。酸度采用酸碱中和滴定法。还原糖采用3, 5-二硝基水杨酸(DNS)法测定[15]。乙醇采用液相色谱仪(Agilent 1200)测定,色谱柱为AminexHPX-87H (300 mm×7.8 mm,Bio-Rad),示差折光检测器(RI)为Schambeck SFD GmbH,测定条件参考Wu等[16]的方法。乳酸采用反相高效液相色谱仪(Waters)测定,色谱柱为Atlantis T3 C18柱(250 mm×4.6 mm),流动相为0.5%的NaH2PO4 (pH 2.7),测定条件:进样量10 μL,流速0.25 mL/min,检测器波长210 nm,柱温40 ℃,保留时间14 min。挥发性化合物的测定采用气相色谱-质谱联仪(GC-MS;Agilent 6890N气相色谱仪和Agilent 5975质谱检测器),色谱柱为DB-Wax (30 m×0.25 mm×0.25 μm;J & W Scientific),内标为薄荷醇(106.25 mg/L),检测条件参考Kong等[17]的报道。
1.3 DNA的提取、PCR扩增和Miseq测序称取7 g样品,根据E.Z.N.A.® soil DNA Kit (Omega Bio-Tek, Norcross,GA,USA)的操作说明提取各样品中的DNA。对于细菌,针对16S的V3-V4区进行扩增,引物为338F (5′-ACTCCTACG GGAGGCAGCAG-3′)和806R (5′-GACTACHVGG GTWTCTAAT-3′)。对于真菌,扩增区域为ITS2区,引物为ITS3 (5′-GCATCGATGAAGAACGCAGC-3′)和ITS4 (5′-TCCTCCGCTTATTGATATGC-3′)。这些引物针对不同的样品都包含有一个8核苷酸barcode的序列。25 μL的反应体系包含2.5 μL的10×Pyrobest buffer,2 μL的dNTPs (2.5 mmol/L),正反向引物各1 μL (10 μmol/L),0.4 μL的Pyrobest DNA polymerase (Takara)和15 ng的模板DNA。细菌的PCR扩增程序:95 ℃预热5 min;95 ℃变性30 s,55 ℃退火30 s,72 ℃延伸40 s,共25个循环,最后72 ℃保持10 min;真菌的PCR扩增程序:95 ℃预热2 min;95 ℃变性30 s,61 ℃退火30 s,72 ℃延伸45 s,共32个循环,最后72 ℃保持10 min。PCR产物根据之前的报道进行纯化[18]。纯化后的产物经NanodropND-1000 UV-Vis定量,然后将PCR产物等分子量混合,并根据Low Sample Protocol进行文库的制备,最后在Illumina Miseq (PE300)平台上机测序。测序后所有的原始序列保存在DDBJ数据库,接受号为DRA005468和DRA005469。
1.4 序列处理原始序列使用QⅡME pipeline进行处理[19]:(1)序列归类,去除标签、引物和接头序列;(2)去除长度小于110 bp,模糊碱基大于2和序列平均质量小于30分的序列;(3) UCHIME去除嵌合体。最后使用UCLUST把高质量的序列根据97%的序列相似度聚成不同的操作性分类单元(OTU),并计算相应的Chao1和Shannon指数。
1.5 数据分析 1.5.1 冗余分析(RDA): 为寻找出环境因子和微生物群落或风味轮廓之间的关系,在R语言中使用Vegan包分析环境因子对群落结构的影响,并进行蒙特卡洛置换检验。 1.5.2 Heatmap绘制和聚类分析: 为找出风味化合物随发酵时间的动态变化规律和样本之间的相似度,在R语言中使用ComplexHeatmap包进行数据标准化、样品和变量的聚类以及热图的绘制。 1.5.3 网络绘制: 计算微生物与代谢物之间的Spearman相关系数(ρ),找出p>0.5且P < 0.05的连接作为可视化对象。对于微生物之间的共现性(co-occurrence)分析,首先计算微生物之间的Spearman相关系数,然后选取|ρ|>0.8且P < 0.05的作为互作对象。使用Cytoscape 3.4.0 (http://www.cytoscape.org)对微生物和代谢物以及微生物与微生物之间的相互关系进行可视化,以表征微生物对代谢物的贡献和微生物与微生物之间的共现性关系。 2 结果和分析 2.1 发酵过程中微生物群落结构组成及其动态变化通过Illumina Miseq测序来确定发酵过程中的微生物群落组成。对于细菌,质控后共得到450273条高质量的序列,平均每个样本具有28142±5001条;并得到3824个OTUs,每个样品共得到239±95个OTUs。对于真菌,质控后共得到450273条高质量的序列,平均每个样本具有89898±39131条;并得到1799个OTUs,每个样品共得到112±15个OTUs。每个样品的覆盖率均在99%以上,表明足够的测序深度和可靠的数据质量[20]。具体的测序信息如Chao1和Shannon指数等见表 1。
| Sample ID | 16S | ITS | |||||||||
| Reads | OTUs | Good’s coverage/% | Chao1 | Shannon | Reads | OTUs | Good’s coverage/% | Chao1 | Shannon | ||
| GN0_1 | 29562 | 199 | 99.86 | 227 | 3.01 | 91193 | 90852 | 99.95 | 125 | 3.32 | |
| GN0_2 | 20254 | 149 | 99.79 | 214 | 2.39 | 93725 | 93287 | 99.95 | 129 | 3.57 | |
| GN5_1 | 31869 | 382 | 99.73 | 450 | 3.08 | 84125 | 83952 | 99.96 | 124 | 3.73 | |
| GN5_2 | 33317 | 306 | 99.74 | 376 | 2.86 | 51580 | 51443 | 99.97 | 104 | 3.11 | |
| GN8_1 | 27029 | 238 | 99.76 | 315 | 2.51 | 146085 | 145843 | 99.95 | 125 | 3.15 | |
| GN8_2 | 31969 | 376 | 99.73 | 451 | 3.03 | 65313 | 65071 | 99.94 | 143 | 3.63 | |
| GN10_1 | 20693 | 315 | 99.47 | 474 | 2.79 | 199146 | 198786 | 99.94 | 134 | 2.34 | |
| GN10_2 | 27161 | 383 | 99.78 | 425 | 3.59 | 99743 | 99414 | 99.95 | 128 | 3.32 | |
| GN15_1 | 27722 | 221 | 99.69 | 329 | 1.17 | 59279 | 59111 | 99.95 | 121 | 3.07 | |
| GN15_2 | 33162 | 167 | 99.78 | 278 | 1.01 | 54972 | 54790 | 99.95 | 116 | 3.00 | |
| GN20_1 | 27095 | 292 | 99.63 | 376 | 1.23 | 64047 | 63805 | 99.96 | 109 | 3.45 | |
| GN20_2 | 34782 | 115 | 99.89 | 161 | 1.26 | 129654 | 129513 | 99.95 | 102 | 2.43 | |
| GN30_1 | 34850 | 233 | 99.70 | 357 | 0.83 | 53366 | 53158 | 99.97 | 105 | 3.12 | |
| GN30_2 | 19903 | 199 | 99.56 | 297 | 1.35 | 91661 | 91576 | 99.96 | 91 | 1.78 | |
| GN40_1 | 25459 | 174 | 99.65 | 325 | 0.89 | 123240 | 122946 | 99.93 | 136 | 2.15 | |
| GN40_2 | 25446 | 75 | 99.88 | 117 | 1.08 | 116432 | 116129 | 99.97 | 106 | 3.28 | |
通过统计发现,在真菌群落中,0–8 d主要由3个真菌目主导,包括Saccharomycetales、Eurotiales和Mucorales;8 d之后,则由Saccharomycetales和Trichosporonales替代(图 1-A)。对于细菌,在最初的10 d内,Lactobacillales和Bacillales占优;之后Lactobacillales迅速增长并占据绝对优势。总体来讲,Saccharomycetales (51.12%–95.21%)和Lactobacillales (39.39%–94.65%)在整个发酵过程中具有很高的优势性。在属水平共得到69个真菌属和215个细菌属。在这些属中,Pichia (11.27%–49.51%)、Dipodascus (4.03%–41.38%)、Candida (1.48%–25.88%)、Saccharomyces (1.19%–13.48%)和Aspergillus (1.19%–13.84%)在整个发酵过程中具有一定的优势。Bacillus、Enterococcus和Lactococcus在0–10 d占优,10 d后Lactobacillus迅速从6.03%增长到92.80% (图 1-B)。

|
| 图 1 发酵过程中微生物群落结构在目水平(A)和属水平(B)的分布变化 Figure 1 Distribution of microbial community at the order level (A) and at the genus level (B) in fermented grains during the fermentation process |
2.2 发酵过程中代谢物的变化
在发酵过程中,共鉴定出61种代谢物,包括5种醇类、10种酸类、25种酯类、17种芳香族化合物和4种杂环类化合物(图 2)。基于这些化合物相对含量的聚类分析表明,样本被分成了2个发酵阶段(阶段Ⅰ:0–20 d;阶段Ⅱ:20–40 d)。代谢物则可以分成3组:组Ⅰ包含21种化合物(9种酯类,6种酸类,5种芳香类和1种杂环类);组Ⅱ包含37种物质(4种酸类,11种芳香类,16种酯类,5种醇类,1种杂环类);组Ⅲ则包含有3种物质,分别是2, 4-二叔丁基苯酚、糠醛和四甲基吡嗪。组Ⅰ的化合物在前期(10–15 d)和后期(20–40 d)均有产生;组Ⅱ中的化合物几乎在后期才产生;组Ⅲ在发酵初始(0 d和5 d)和发酵后期(20 d之后)的含量相对较高。这表明20 d之后的发酵期是白酒发酵过程中的风味物质高产期。

|
| 图 2 酒醅发酵过程中风味化合物的变化热图 Figure 2 Heatmap of change in metabolic profiling during the fermentation process. Clustering analysis was performed using Pearson correlation and Euclidean distance based on the relative content of metabolites during the fermentation process. Data were processed by z-score transformation |
2.3 揭示酿造核心微生物群
我们选取丰度在前20的细菌属和前20的真菌属,计算这40个属与61种代谢物之间的Spearman相关系数(p),选取p>0.5且P < 0.05作为有效的网络连接并绘图如图 3。可以发现,Lactobacillus和Saccharomyces是连接数最大的2个属(36和22),相关的化合物主要是酯类、酸类、醇类和芳香族化合物,尤其是这2个属都贡献重要物质乙酸(VC10)和乙醇(E),且Lactobacillus与乳酸(VC61)也有显著的正相关。Candida与辛酸乙酯(VC44),乙酸异戊酯(VC3)、2-甲基-1-丁醇(VC1)、苯乙醇(VC54)、异戊醇(VC2)和乙醇(E)都有一定的正相关性。还原糖(S)和糠醛(VC24)则与多个属具有正向的联系。其次是Dekkera、Pseudeurotium、Trichosporon与一些酯类、芳香类和杂环类化合物具有显著的相关性。以上结果表明在白酒酒醅发酵过程中,比较重要的代谢产物贡献者是Lactobacillus、Saccharomyces和Candida。具体的网络图信息见表 1。

|
| 图 3 微生物和风味化合物的相关性网络图 Figure 3 Correlation network between microbial genera (pink) and compounds (other colored circles indicate different groups of compounds) calculated by statistically significant (P < 0.05) Spearman correlation coefficient (p>0.5). The size of nodes indicates the degree of connections. Edge thickness represents the proportional to the value of Spearman's correlation |
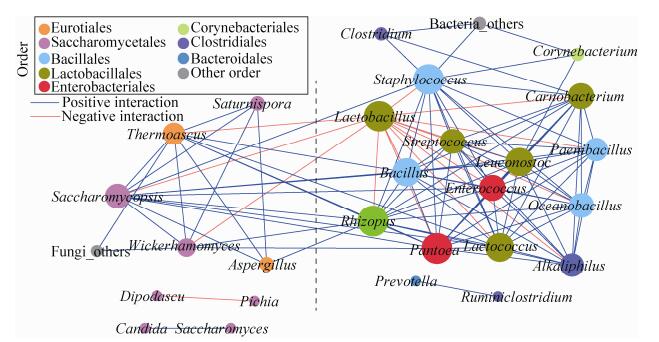
为探究微生物之间的相互作用关系,通过共现性网络分析(co-occurrence network analysis),共得到38个有效的连接节点(nodes)和267个边(edges)。网络的聚类系数为0.622,较强的连接节点(每个节点有超过12条边的连接)大多分布在Lactobacillales、Enterobacteriales和Bacillales,包括Lactobacillus、Bacillus、Enterococcus、Lactococcus、Pantoea、Rhizopus、Carnobacterium、Acinetobacter、Leuconostoc、Oceanobacillus、Paenibacillus、Saccharomycopsis、Staphylococcus、Streptococcus、Alkaliphilus、Thermoascus、Clostridium、Wickerhamomyces、Penicillium、Corynebacterium和Trichomonascus,各菌属的共现率(incidence of co-occurrence)在2%以上(图 4),尤其是Lactobacillus与其他15个属具有显著的负相关性。此外,Lactobacillus、Rhizopus、Bacillus、Lactococcus、Saccharomycopsis、Alkaliphilus、Thermoascus、Enterococcus和Pantoea的平均相对丰度在1%以上,且共现率均在3.5%以上。Pichia和Dipodascus的平均相对丰度尤其较高,分别为28.91%和25.95% (表 2)。Alkaliphilus、Enterococcus和Pantoea被认为是来自于动物肠道、植物根部或土壤且对食品发酵具有污染性的菌属[21-23]。
|
| 图 4 酒醅中微生物之间的共现性网络图 Figure 4 Correlation network of co-occurring microbial genera in fermented grains. Statistically significant (P < 0.05) Spearman correlation coefficient (|ρ|>0.8) indicates the robust correlations. The size of nodes indicates the degree of connections. The same color of nodes indicates the same order. Edge thickness represents the proportional to the value of Spearman's correlation. Blue and red edge indicate positive and negative interaction between nodes |
| Genus | Average abundance/% | Highest abundance/% | Lowest abundance/% | Incidence of co-occurrence/% |
| Lactobacillus | 52.53 | 93.27 | 3.15 | 4.68 |
| Bacillus | 16.78 | 47.23 | 0.88 | 4.49 |
| Enterococcus | 7.55 | 20.19 | 0.39 | 4.49 |
| Lactococcus | 6.43 | 16.48 | 0.20 | 4.49 |
| Rhizopus | 4.71 | 24.83 | 0.03 | 4.49 |
| Pantoea | 2.15 | 4.97 | 0.16 | 4.49 |
| Saccharomycopsis | 1.86 | 10.80 | 0.08 | 4.12 |
| Alkaliphilus | 1.35 | 3.77 | 0.07 | 3.75 |
| Thermoascus | 1.14 | 5.57 | 0.04 | 3.75 |
| Saccharomyces | 9.73 | 13.84 | 5.87 | 1.31 |
| Candida | 18.19 | 25.88 | 7.92 | 0.75 |
| Dipodascus | 25.95 | 41.38 | 4.03 | 0.19 |
| Pichia | 28.91 | 49.51 | 11.27 | 0.19 |
| Incidence of co-occurrence is the ratio of targeted edges to total edges. | ||||
根据风味贡献程度,相互作用关系强度和相对丰度(图 3,图 4,表 2),得到发酵过程中的核心微生物群共包含10个属:Lactobacillus、Saccharomyces、Candida、Rhizopus、Saccharomycopsis、Pichia、Dipodascus、Bacillus、Thermoascus和Lactococcus。为验证所鉴定的核心微生物群是否推动了风味的演变,通过提取二者的第一主成分并进行相关性分析后发现,核心微生物群与风味轮廓的演变具有极显著的相关性(图 5,r2=0.627,P < 0.001),说明这10个属所组成的核心微生物群是白酒发酵过程中风味演替的主要推动者。

|
| 图 5 微生物群落结构和风味结构的相关性 Figure 5 Correlations between core microbiota PC1 (56.74%) and metabolic PC1 (45.17%) |
2.4 环境因素对核心微生物群的影响
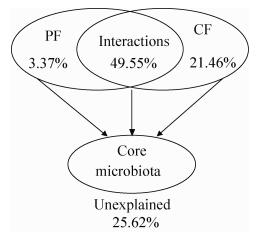
为揭示由10个属组成的核心微生物群和环境因子之间的关系,使用冗余分析进行定量化分析并进行蒙特卡洛置换检验。首先将环境因子分为两类:物理因素包括水分、酸度和温度,化学因素包括还原糖、乙醇、乳酸和乙酸(表 2)。冗余分析表明7个因素能够解释绝大部分的核心微生物群的动态变化(74.38%),说明环境因素的确是驱动群落演变的主要因素(图 6)。蒙特卡洛置换检验的结果表明,对核心微生物群的演替具有重要影响的因素共有5个:水分、酸度、温度、还原糖和乙醇,因为这5个因素与该微生物群的分布具有显著的相关性(r2>0.5,P < 0.01) (表 3)。化学因素对群落变化的解释性(21.46%)要高于物理因素(3.37%) (图 6),表明核心微生物群的演变主要是由微生物产生的代谢物引起的化学因素所驱动的。此外,物理因素和化学因素的互作对其演变也具有非常重要的影响(49.55%)。如表 4所示,还原糖几乎与除Lactobacillus和Saccharomyces外的所有菌属具有显著的正相关;其他6个环境因素则与Lactobacillus、Candida和Saccharomyces具有一定的正相关性,而与Bacillus、Lactococcus、Rhizopus、Thermoascus和Saccharomycopsis具有一定的负相关性。
|
| 图 6 环境因素对核心微生物群的影响及其解释性 Figure 6 Variation partitioning of the core microbiota among physical factors (PF: moisture, acidity and temperature), chemical factors (CF: reducing sugar, ethanol, lactic acid and acetic acid) and their interactions |
| Monte Carlo permutation test | r2 | P |
| Moisture/% | 0.7172** | 0.003 |
| Acidity/(g/100g) | 0.5273** | 0.009 |
| Temperature/℃ | 0.7016*** | 0.001 |
| Reducing sugar/(mg/g) | 0.6770*** | 0.001 |
| Ethanol/(mg/g) | 0.9042*** | 0.001 |
| Lactic acid/(mg/g) | 0.2435 | 0.164 |
| Acetic acid/(mg/g) | 0.3558 | 0.069 |
| Genus | Moisture/% | Acidity/ (g/100g) |
T/℃ | Reducing sugar/ (mg/g) |
Ethanol/ (mg/g) |
Lactic acid/ (mg/g) |
Acetic acid/ (mg/g) |
| Lactobacillus | 0.696** | 0.644** | 0.736** | –0.929** | 0.909** | 0.732** | 0.615* |
| Bacillus | –0.726** | –0.515* | –0.809** | 0.856** | –0.906** | –0.718** | –0.662** |
| Lactococcus | –0.692** | –0.391 | –0.757** | 0.885** | –0.847** | –0.679** | –0.609* |
| Pichia | –0.090 | –0.035 | –0.149 | 0.106 | –0.285 | –0.341 | –0.409 |
| Dipodascus | 0.147 | 0.179 | 0.035 | 0.006 | 0.147 | 0.232 | 0.176 |
| Candida | 0.405 | 0.571* | 0.493 | –0.353 | 0.594* | 0.162 | 0.412 |
| Saccharomyces | 0.642** | 0.479 | 0.557* | –0.512* | 0.603* | 0.300 | 0.550* |
| Rhizopus | –0.712** | –0.635** | –0.758** | 0.879** | –0.912* | –0.721** | –0.550* |
| Saccharomycopsis | –0.690** | –0.521* | –0.672** | 0.803** | –0.812** | –0.609* | –0.482 |
| Thermoascus | –0.655** | –0.600* | –0.784** | 0.782** | –0.835** | –0.597* | –0.350 |
| Red and green indicate positive and negative correlation with each environmental factors. The bold indicates significant differences. **, P < 0.01; *, P < 0.05. | |||||||
3 讨论
近年来,有关发酵食品的研究越来越多地集中在微生物群落结构的挖掘上。据报道,许多分子手段尤其是二代测序技术已经广泛应用到食品发酵的研究当中,为我们了解食品发酵的本质提供了更广阔的视角[24]。由于大曲和新粮的加入以及酒醅暴露在空气中,在白酒的固态发酵前期,在酒醅入窖池时,会有多种微生物混入其中,从而导致前期的微生物群落结构比较离散。随着发酵的进行,由于温度、酸度和乙醇的升高以及可利用还原糖的下降,窖池内的生态环境逐渐变成一个相对极端的环境,导致后期发酵微生物群落尤其是细菌群落的相对集中。
不同的发酵食品由不同的核心微生物组成,如Acetobactor和Lactobacillus对于醋的发酵[25],Lactobacillus和Streptococcus对于奶酪的发酵[26]。核心微生物群对于保证发酵食品的风味多样性和产品质量的批次稳定性具有重要的作用[27]。本研究从白酒发酵过程中共揭示出10个核心微生物属。Saccharomyces是白酒酿造过程中的优势酵母属之一[16],且能够产生多种风味酯并贡献乙醇和其他有机酸[28]。Lactobacillus是该过程的优势细菌菌属,如Lactobacillus rhamnosu、Lactobacillus fermentum、Lactobacillus acetotolerans、Lactobacillus kefiri等乳酸菌在不同的发酵食品中能够贡献乙醇、乙酸、乳酸和其他的重要风味物质[29-31],由于这些物质(乙醇、乙酸和乳酸等)能够较快地改变发酵过程中的生态环境,使得其他不耐受微生物的生长受到抑制,这也是Lacobacillus与很多其他菌属具有负相关,并且使得化学因素成为驱动菌群变化的重要原因。有报道指出,Lactococcus具有一定的氨肽酶活性和蛋白质水解力,并且有一定的风味贡献功能[32]。Bacillus、Candida、Pichia、Saccharomycopsis以及Rhizopus是大曲成熟过程中的优势菌属之一[33],前三者在白酒酿造中具有潜在的风味贡献功能[34-35],如 Bacillus能够产生多种酚类和含氮类杂环化合物。Saccharomycopsis和Rhizopus则具有较高的淀粉酶和糖苷酶活性[36],Thermoascus能够产生热稳定的水解酶如纤维素酶和木聚糖酶[37],这些菌属在白酒酿造过程中能够降解谷物中的淀粉或纤维素,是白酒发酵不可或缺的重要菌属。而目前关于Dipodascus在发酵食品中的报道较少,但在本研究中由于所检测到的比例较高,所以也将其列为核心微生物属之一。以上10个属所组成的核心微生物群与风味的演变具有显著的相关性,说明这10个属组成的核心微生物群是白酒发酵过程中的关键微生物群。
本研究从微生物组和代谢组的角度,结合多元统计学分析更加全面地揭示了白酒发酵过程中的核心微生物群,并量化了各环境因素对核心微生物群的影响,为下一步白酒酿造的定向调控提供参考。
| [1] | Giraffa G. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiology Reviews, 2004, 28(2): 251-260. DOI:10.1016/j.femsre.2003.10.005 |
| [2] | Smid EJ, Lacroix C. Microbe-microbe interactions in mixed culture food fermentations. Current Opinion in Biotechnology, 2013, 24(2): 148-154. DOI:10.1016/j.copbio.2012.11.007 |
| [3] | Großkopf T, Soyer OS. Synthetic microbial communities. Current Opinion in Microbiology, 2014, 18: 72-77. DOI:10.1016/j.mib.2014.02.002 |
| [4] | Wolfe BE, Dutton RJ. Fermented foods as experimentally tractable microbial ecosystems. Cell, 2015, 161(1): 49-55. DOI:10.1016/j.cell.2015.02.034 |
| [5] | De Roy K, Marzorati M, van den Abbeele P, van de Wiele T, Boon N. Synthetic microbial ecosystems:an exciting tool to understand and apply microbial communities. Environmental Microbiology, 2014, 16(6): 1472-1481. DOI:10.1111/emi.2014.16.issue-6 |
| [6] | Wolfe BE, Button JE, Santarelli M, Dutton RJ. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell, 2014, 158(2): 422-433. DOI:10.1016/j.cell.2014.05.041 |
| [7] | de Pasquale I, di Cagno R, Buchin S, de Angelis M, Gobbetti M. Microbial ecology dynamics reveal a succession in the core microbiota involved in the ripening of pasta filata caciocavallo pugliese cheese. Applied and Environmental Microbiology, 2014, 80(19): 6243-6255. DOI:10.1128/AEM.02097-14 |
| [8] | Wang ZM, Lu ZM, Shi JS, Xu ZH. Exploring flavour-producing core microbiota in multispecies solid-state fermentation of traditional Chinese vinegar. Scientific Reports, 2016, 6: 26818. DOI:10.1038/srep26818 |
| [9] | Hu XL, Du H, Ren C, Xu Y. Illuminating anaerobic microbial community and cooccurrence patterns across a quality gradient in chinese liquor fermentation pit muds. Applied and Environmental Microbiology, 2016, 82(8): 2506-2515. DOI:10.1128/AEM.03409-15 |
| [10] | Rui JP, Li JB, Zhang SH, Yan XF, Wang YP, Li XZ. The core populations and co-occurrence patterns of prokaryotic communities in household biogas digesters. Biotechnology for Biofuels, 2015, 8: 158. DOI:10.1186/s13068-015-0339-3 |
| [11] | Chaillou S, Chaulot-Talmon A, Caekebeke H, Cardinal M, Christieans S, Denis C, Desmonts MH, Dousset X, Feurer C, Hamon E, Joffraud JJ, La Carbona S, Leroi F, Leroy S, Lorre S, Macé S, Pilet MF, Prévost H, Rivollier M, Roux D, Talon R, Zagorec M, Champomier-Vergès MC. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. The ISME Journal, 2015, 9(5): 1105-1118. DOI:10.1038/ismej.2014.202 |
| [12] | Cardinale M, Grube M, Erlacher A, Quehenberger J, Berg G. Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environmental Microbiology, 2015, 17(1): 239-252. DOI:10.1111/1462-2920.12686 |
| [13] | Parente E, Cocolin L, de Filippis F, Zotta T, Ferrocino I, O'Sullivan O, Neviani E, de Angelis M, Cotter PD, Ercolini D. Food Microbionet:A database for the visualisation and exploration of food bacterial communities based on network analysis. International Journal of Food Microbiology, 2016, 219: 28-37. DOI:10.1016/j.ijfoodmicro.2015.12.001 |
| [14] | Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. The ISME Journal, 2012, 6(2): 343-351. DOI:10.1038/ismej.2011.119 |
| [15] | Wang Y, Yuan B, Ji YC, Li H. Hydrolysis of hemicellulose to produce fermentable monosaccharides by plasma acid. Carbohydrate Polymers, 2013, 97(2): 518-522. DOI:10.1016/j.carbpol.2013.05.017 |
| [16] | Wu Q, Chen LQ, Xu Y. Yeast community associated with the solid state fermentation of traditional Chinese Maotai-flavor liquor. International Journal of Food Microbiology, 2013, 166(2): 323-330. DOI:10.1016/j.ijfoodmicro.2013.07.003 |
| [17] | Kong Y, Wu Q, Zhang Y, Xu Y. In situ analysis of metabolic characteristics reveals the key yeast in the spontaneous and solid-state fermentation process of Chinese light-style liquor. Applied and Environmental Microbiology, 2014, 80(12): 3667-3376. DOI:10.1128/AEM.04219-13 |
| [18] | Ren GD, Ren WJ, Teng Y, Li ZG. Evident bacterial community changes but only slight degradation when polluted with pyrene in a red soil. Frontiers in Microbiology, 2015, 6: 22. |
| [19] | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QⅡME allows analysis of high-throughput community sequencing data. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [20] | Lemos LN, Fulthorpe RR, Triplett EW, Roesch LFW. Rethinking microbial diversity analysis in the high throughput sequencing era. Journal of Microbiological Methods, 2011, 86(1): 42-51. DOI:10.1016/j.mimet.2011.03.014 |
| [21] | Gobbetti M, Minervini F, Pontonio E, di Cagno R, de Angelis M. Drivers for the establishment and composition of the sourdough lactic acid bacteria biota. International Journal of Food Microbiology, 2016, 239: 3-18. DOI:10.1016/j.ijfoodmicro.2016.05.022 |
| [22] | Ambardar S, Sangwan N, Manjula A, Rajendhran J, Gunasekaran P, Lal R, Vakhlu J. Identification of bacteria associated with underground parts of Crocus sativus by 16S rRNA gene targeted metagenomic approach. World Journal of Microbiology & Biotechnology, 2014, 30(10): 2701-2709. |
| [23] | Cai L, Yu K, Yang Y, Chen BW, Li XD, Zhang T. Metagenomic exploration reveals high levels of microbial arsenic metabolism genes in activated sludge and coastal sediments. Applied Microbiology and Biotechnology, 2013, 97(21): 9579-9588. DOI:10.1007/s00253-012-4678-8 |
| [24] | Kergourlay G, Taminiau B, Daube G, Vergès MCC. Metagenomic insights into the dynamics of microbial communities in food. International Journal of Food Microbiology, 2015, 213: 31-39. DOI:10.1016/j.ijfoodmicro.2015.09.010 |
| [25] | Li S, Li P, Liu X, Luo LX, Lin WF. Bacterial dynamics and metabolite changes in solid-state acetic acid fermentation of Shanxi aged vinegar. Applied Microbiology and Biotechnology, 2016, 100(10): 4395-4411. DOI:10.1007/s00253-016-7284-3 |
| [26] | Monnet C, Dugat-Bony E, Swennen D, Beckerich JM, Irlinger F, Fraud S, Bonnarme P. Investigation of the activity of the microorganisms in a reblochon-style cheese by metatranscriptomic analysis. Frontiers in Microbiology, 2016, 7: 536. |
| [27] | Awasthi A, Singh M, Soni SK, Singh R, Kalra A. Biodiversity acts as insurance of productivity of bacterial communities under abiotic perturbations. The ISME Journal, 2014, 8(12): 2445-2452. DOI:10.1038/ismej.2014.91 |
| [28] | Wu Q, Xu Y, Chen LQ. Diversity of yeast species during fermentative process contributing to Chinese Maotai-flavour liquor making. Letters in Applied Microbiology, 2012, 55(4): 301-307. DOI:10.1111/lam.2012.55.issue-4 |
| [29] | Laakso K, Koskenniemi K, Koponen J, Kankainen M, Surakka A, Salusjärvi T, Auvinen P, Savijoki K, Nyman TA, Kalkkinen N, Tynkkynen S, Varmanen P. Growth phase-associated changes in the proteome and transcriptome of Lactobacillus rhamnosus GG in industrial-type whey medium. Microbial Biotechnology, 2011, 4(6): 746-766. DOI:10.1111/mbt.2011.4.issue-6 |
| [30] | Annan NT, Poll L, Sefa-Dedeh S, Plahar WA, Jakobsen M. Volatile compounds produced by Lactobacillus fermentum, Saccharomyces cerevisiae and Candida krusei in single starter culture fermentations of Ghanaian maize dough. Journal of Applied Microbiology, 2003, 94(3): 462-474. DOI:10.1046/j.1365-2672.2003.01852.x |
| [31] | de Angelis M, Calasso M, Cavallo N, di Cagno R, Gobbetti M. Functional proteomics within the genus Lactobacillus. Proteomics, 2016, 16(6): 946-962. DOI:10.1002/pmic.v16.6 |
| [32] | Centeno JA, Tomillo FJ, Fernández-García E, Gaya P, Nuñez M. Effect of wild strains of Lactococcus lactis on the volatile profile and the sensory characteristics of ewes' raw milk cheese. Journal of Dairy Science, 2002, 85(12): 3164-3172. DOI:10.3168/jds.S0022-0302(02)74404-4 |
| [33] | Liu X, Guo KL, Zhang HX. Determination of microbial diversity in Daqu, a fermentation starter culture of Maotai liquor, using nested PCR-denaturing gradient gel electrophoresis. World Journal of Microbiology & Biotechnology, 2012, 28(6): 2375-2381. |
| [34] | Rojas V, Gil JV, Piñaga F, Manzanares P. Studies on acetate ester production by non-Saccharomyces wine yeasts. International Journal of Food Microbiology, 2001, 70(3): 283-289. DOI:10.1016/S0168-1605(01)00552-9 |
| [35] | Meng X, Wu Q, Wang L, Wang DQ, Chen LQ, Xu Y. Improving flavor metabolism of Saccharomyces cerevisiae by mixed culture with Bacillus licheniformis for Chinese Maotai-flavor liquor making. Journal of Industrial Microbiology & Biotechnology, 2015, 42(12): 1601-1608. |
| [36] | Zheng XW. Daqu-a traditional fermentation starter in China:microbial ecology and functionality. Doctor Dissertation of Wageningen University, 2015. |
| [37] | Jain KK, Dey TB, Kumar S, Kuhad RC. Production of thermostable hydrolases (cellulases and xylanase) from Thermoascus aurantiacus RCKK:a potential fungus. Bioprocess and Biosystems Engineering, 2015, 38(4): 787-796. DOI:10.1007/s00449-014-1320-4 |
 2018, Vol. 58
2018, Vol. 58