中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 冯丽, 蒋群, 孙伟, 李英新, 张风丽, 李志勇. 2017
- Li Feng, Qun Jiang, Wei Sun, Yingxin Li, Fengli Zhang, Zhiyong Li. 2017
- 中国南海部分深海沉积物真菌多样性及其抗菌活性
- Diversity and antimicrobial activity of fungi isolated from deep-sea sediments in South China Sea
- 微生物学报, 57(9): 1332-1341
- Acta Microbiologica Sinica, 57(9): 1332-1341
-
文章历史
- 收稿日期:2017-03-29
- 修回日期:2017-04-24
- 网络出版日期:2017-05-02
高压、低温、寡营养等极端条件使深海成为一种特殊生境,部分深海来源的真菌能产一些有特殊活性的化合物,因此深海真菌被认为是发现新的生物活性物质的重要来源。第1次在深海中发现真菌可追溯到1964年,是在大西洋4450 m水深处[1]。近年来相继有文献报道在一些深海环境中发现了真菌的存在,例如:南海沉积物[2-3]、太平洋沉积物[4-5]、中印度洋和东印度洋海沉积物[6-10]、北极地沉积物[11]等。尽管深海真菌越来越受关注,但是相比细菌和古菌,我们对真菌还知之甚少。尤其是对南海深海沉积物的真菌多样性及其特性了解还比较欠缺,只有少数文献对南海沉积物真菌多样性进行了报道[2-3, 12]。从南海深海沉积物来源的可培养真菌中已分离到了多种具有生物活性的次级代谢产物,例如:邻氨基苯甲酸衍生物[13]、哌啶二酮[14]、黄色素[15]、环戊烯酮[16]、倍半萜[17]、生物碱[18-19]、吲哚二酮哌嗪[20]、细胞松弛素[21]、异戊烯酮类[22]等。由此可见,南海深海沉积物来源的真菌有产生物活性物质的潜能,值得深入研究。
本研究目的是揭示南海深海沉积物可培养真菌的多样性,并比较不同位点和深度之间的差异,同时调查南海深海沉积物可培养真菌产抗菌活性物质的潜能,为进一步利用南海深海真菌奠定基础。
1 材料和方法 1.1 材料 1.1.1 沉积物材料来源: 实验所用的沉积物样本由上海交通大学王风平教授提供,来自南海的4个站位,采样水深分别为1183 m(A)、897 m(B)、约1000 m(C)和1434 m(D),其中A、B站位位于神狐海域,C、D站位位于琼东南盆地。从深海获得的样品直接送到船上分装,每个站位再取3个不同深度,分别来自沉积物柱的大约0.05 m (表层)、1.00 m和2.00 m处,总计12份沉积物样品。样品立即存放在无菌袋中并封口以避免污染,保存于4 ℃,低温运输至实验室后用于真菌的分离。 1.1.2 指示菌种类: 指示菌包括枯草芽孢杆菌(Bacillus subtilis, G+)、蕈状芽孢杆菌(Bacillus mycoides, G+)、金黄色葡萄球菌(Staphylococcus aureus, G+)、鲍曼不动杆菌(Acinetobacter baumanii, G–)、大肠杆菌(Escherichia coli, G–)、铜绿假单胞菌(Pseudomonas aeruginosa, G–)共6株细菌以及1株水稻纹枯病真菌(Rhizotonia solani)。 1.2 真菌分离培养为了尽可能全面揭示沉积物中的可培养真菌多样性,本实验使用了4种不同的方法来分离培养真菌[3, 5, 23-27]。(1) 稀释涂平板法:用灭菌小铲取1 g沉积物样本,置于9 mL灭菌的人工海水中,振荡10 min即得10–1稀释液。无菌吸管吸取1 mL 10–1稀释液加到9 mL灭菌的人工海水中,充分摇匀,即得10–2稀释液。取10–1和10–2稀释液各100 μL涂平板,同时设置2个温度(15 ℃和25 ℃)培养;(2) 低温诱导法:取100 μL 10–1稀释的沉积物涂平板,将平板置于4 ℃培养;(3) 颗粒平板法:采用能通过200 μm网筛但不能通过100 μm网筛的沉积物颗粒涂平板分离真菌;(4) 高通量筛选[28]:也叫极限稀释法,利用48孔板,根据稀释涂平板的结果来确认稀释倍数,确保每个孔中的真菌数不会多于1个。此外共使用了8种培养基分离真菌,所有的培养基成分均采用常规用量的1/5 (表 1),以此来模拟深海的寡营养状态。根据前两种方法得到的结果,颗粒平板法和高通量筛选法只选用了分离效果较好的CYA培养基。所有培养基中均添加青霉素及链霉素(1 g/L)抑制细菌的生长。在分离真菌时,设置空白平板,所有的平板都设3个平行重复,每隔3–5 d观察培养效果,所有长出的真菌菌落均接种于PDA平板上多次纯化直至获得纯的菌株。
| Media | Components of media | Isolates | ||||
| Dilution plating method | Low temperature incubation method | Particle plating method | High-throughput culturing method | Total | ||
| PDA | PDB powder 30 g, Agar 15 g, ASW 1000 mL | 10 | 5 | – | – | 15 |
| CDA | NaNO33 g, K2HPO41 g, MgSO4·7H2O 0.5 g, KCl 0.5 g, FeSO4·7H2O 0.001 g, Agar 15 g, ASW1000 mL | 12 | 5 | – | – | 17 |
| CYA | Czapek’s concentrated solution 2 mL, K2HPO4 1 g, yeast extract 5 g, Agar 15 g, ASW 1000 mL | 13 | 6 | 12 | 10 | 19+22 |
| MEA | Malt extract 30 g, Peptone 3 g, Agar 15 g, ASW 1000 mL | 10 | 4 | – | – | 14 |
| SA | Peptone 10 g, Glucose 40 g, Agar 15 g, ASW 1000 mL | 9 | 4 | – | – | 13 |
| CMA | Corn extract 7 g, Agar 15 g, ASW 1000 mL | 5 | 2 | – | – | 7 |
| GYPS | Glucose 5 g, Yeast extract 5 g, Starch 5 g, Agar 15 g, ASW 1000 mL | 5 | 3 | – | – | 8 |
| BSA | Malt extract 15 g, Yeast extract 2 g, Benomyl 10 μg, Lactic acid 2 mL, Agar 15 g, Chloramphenicol 50 μg, ASW 1000 mL | 6 | 4 | – | – | 10 |
| ASW: Abbreviation of artificial sea water. All the above media were used at 1/5 strength. –: The media were not used. | ||||||
1.2.1 真菌鉴定: 对纯培养的125株真菌采用CTAB法提取DNA,使用引物ITS1 (5'-TCCGTAG GTGAACCTGCGG-3')和ITS4 (5'-TCCTCCGCTT ATTGATATGC-3')扩增ITS序列进行分子鉴定。将所测得的序列在NCBI数据库中进行BLAST比对,结合形态学分析和序列比对对真菌进行排重[29],然后基于MEGA软件中的邻接法(NJ)进行系统发育分析。 1.2.2 真菌培养与浸膏制备: 挑取经活化的真菌接种于改良查氏液体培养基中,于25 ℃、120 r/min培养10 d。5000 r/min离心20 min,收集菌丝体和发酵液。菌丝体先经超声破碎,再经乙酸乙酯浸泡,发酵液用乙酸乙酯等比萃取,二者混合后蒸干,提取物用乙酸乙酯溶解成5 mg/mL的粗浸膏备用。 1.2.3 抑菌活性分析: 可培养真菌发酵液粗浸膏的抑细菌活性采用滤纸片扩散法[30]。将保存于甘油管中的指示细菌,接种到新鲜的营养琼脂平板上活化,37 ℃培养18 h。将平板上的菌落接种到锥形瓶中培养一段时间后,将菌悬液浓度调整为107 CFU/ mL。取100 μL菌悬液在营养琼脂平板上均匀涂布。将20 μL粗浸膏分多次滴加于滤纸片,待乙酸乙酯挥发后,置于涂有指示菌的平板,37 ℃培养12 h,以抑菌圈的直径来表示粗浸膏抑菌活性的强弱。每个样本设置3个平行重复,同时设置一个阴性对照(乙酸乙酯挥发后的空白滤纸片),一个阳性对照(G+以氨苄青霉素作为阳性对照,G–以链霉素作为阳性对照)。采用生长速率法测定抑真菌活性[31]。在无菌条件下,取制备好的粗浸膏1 mL,分别加入到培养皿中,再加入15 mL冷却至40 ℃左右的PDA培养基充分混匀,冷却凝固后备用。将真菌指示菌打成大小一致的菌饼,接种于平板上。以含有等量乙酸乙酯的培养基平板作为对照,28 ℃、3 d后测定生长直径,以指示真菌菌落直径差来表示抑菌活性强弱。 2 结果和分析 2.1 可培养真菌的多样性
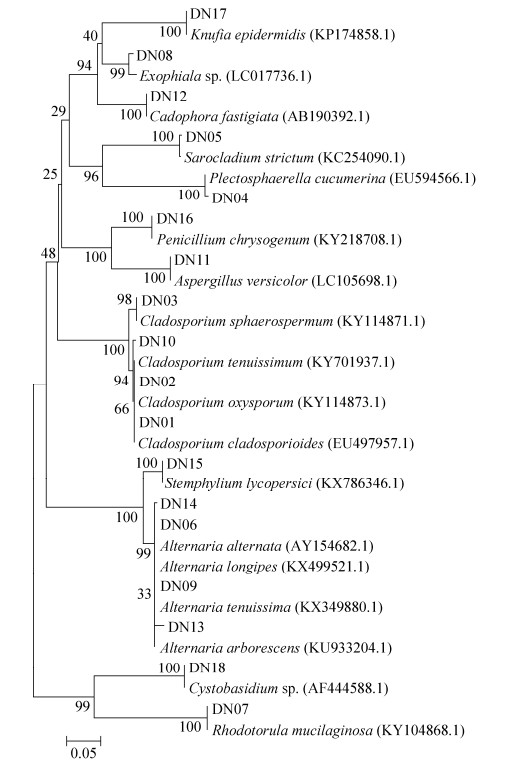
总共从12份沉积物中分离出125株纯培养的真菌(表 2),形态学结合ITS序列比对排重后得到18个种类型,其GenBank登录号为KY781369– KY781386(DN01–DN18)。基于ITS序列比对分析,这些菌株可归属于子囊菌门(Ascomycota)和担子菌门(Basidiomycota)的12个属。可培养真菌在4个站位的多样性与分布见表 2,NJ法构建的系统发育树见图 1。可培养真菌的ITS序列比对结果表明,所有的序列与数据库中已有序列的相似度都 > 97%。可培养的真菌大多数属于子囊菌门,只有2株真菌是属于担子菌门的酵母(DN07和DN18)。所有分离到的真菌属于6个纲、12个属,这其中枝孢属(Cladosporium)是最优势的属,其次是链格孢属(Alternaria)。
| Isolates | Closest relative (GenBank No.) | Class | Similarity/% | A | B | C | D | |||||||||||
| A-0 | A-1 | A-2 | B-0 | B-1 | B-2 | C-0 | C-1 | C-2 | D-0 | D-1 | D-2 | |||||||
| DN01 | Cladosporium cladosporioides (EU497957.1) | Dothideomycetes | 100 | ● | ● | ● | ||||||||||||
| DN02 | Cladosporium oxysporum (KY114873.1) | Dothideomycetes | 100 | ● | ● | |||||||||||||
| DN03 | Cladosporium sphaerospermum (KY114871.1) | Dothideomycetes | 100 | ● | ● | ● | ● | ● | ● | ● | ||||||||
| DN04 | Plectosphaerella cucumerina (EU594566.1) | Sordariomycetes | 100 | ● | ||||||||||||||
| DN05 | Sarocladium strictum (KC254090.1) | Sordariomycetes | 100 | ● | ● | ● | ||||||||||||
| DN06 | Alternaria arborescens (KU933204.1) | Dothideomycetes | 99 | ● | ● | ● | ||||||||||||
| DN07 | Rhodotorula mucilaginosa (KY104868.1) | Microbotryomycetes | 99 | ● | ||||||||||||||
| DN08 | Exophiala sp. (LC017736.1) | Eurotiomycetes | 99 | ● | ● | ● | ● | |||||||||||
| DN09 | Alternaria tenuissima (KX349880.1) | Dothideomycetes | 100 | ● | ||||||||||||||
| DN10 | Cladosporium tenuissimum (KP701937.1) | Dothideomycetes | 99 | ● | ● | ● | ||||||||||||
| DN11 | Aspergillus versicolor (LC105698.1) | Eurotiomycetes | 100 | ● | ||||||||||||||
| DN12 | Cadophora fastigiata (AB190392.1) | Leotiomycetes | 100 | ● | ||||||||||||||
| DN13 | Alternaria longipes (KX499521.1) | Dothideomycetes | 99 | ● | ● | |||||||||||||
| DN14 | Alternaria alternata (AY154682.1) | Dothideomycetes | 99 | ● | ||||||||||||||
| DN15 | Stemphylium lycopersici (KX786346.1) | Dothideomycetes | 100 | ● | ||||||||||||||
| DN16 | Penicillium chrysogenum (KY218708.1) | Eurotiomycetes | 100 | ● | ● | |||||||||||||
| DN17 | Knufia epidermidis (KP174858.1) | Eurotiomycetes | 99 | ● | ||||||||||||||
| DN18 | Cystobasidium sp. (AF444588.1) | Cystobasidiomycetes | 100 | ● | ||||||||||||||
| A-0: Site A, the deepth is about 0.05 m; A-1: Site A, the deepth is about 1.00 m; A-2: Site A, The deepth is about 2.00 m. Other sites were the same. ●: The fungi was isolated from this sample. | ||||||||||||||||||
|
| 图 1 基于NJ法对18株代表性真菌的ITS rDNA序列构建系统发育树 Figure 1 Neighbour-Joining (NJ) tree based on fungal ITS rDNA sequences of 18 fungal isolates. 0.05 was sequence divergence. |
对4个站位分离培养的真菌进行比较,Cladosporium sp.和Alternaria sp.在4个站位中都有分离到,Exophiala sp.只在B、D站位分离到,其他属都只在一个站位分离到。进一步使用Bray-Curtis来计算每个站位之间的差异度,发现B和C两个位点的差异度为100%,其他各站位两两之间的差异度在70%–90%之间,这说明4个站位之间的可培养真菌多样性差异较大。2株担子菌门的酵母(DN07和DN18) 均来自最底部样品(即约2 mbsf (below the seafloor)处)。A站位一共分到了7种真菌,其中A-2深度分离到最多种类的真菌;枝孢属和帚枝霉属(Sarocladium)为优势属真菌,在3个深度都存在;B站位一共分到3种真菌,B-0和B-2处都分到2种,外瓶霉属(Exophiala)在3个深度的样品中都存在;C站位一共得到6种真菌,C-2分到最多,枝孢属和链格孢属的真菌在3个深度样品都存在;D站位一共分到8种真菌,3个深度样品各分到3种真菌,青霉属(Penicillium)在D-0和D-1都分离到。使用方差分析计算每个站位中深度对真菌多样性的影响,得到的P值均大于0.05,说明1–2 m的样品采样深度差异对真菌多样性没有显著的影响。
2.2 抑菌活性选择18种代表性可培养真菌进行液体发酵制备粗浸膏,进行抑菌活性筛选,发现10株真菌对6株细菌指示菌有不同程度的抑制效果(表 3)。这10株真菌都属于子囊菌门。其中Stemphylium lycopersici DN15对3种G+细菌都有较好的抑菌效果;Alternaria alternata DN14对2种G+细菌有抑制作用,而且对G–铜绿假单胞菌也有抑制作用;菌株Penicillium chrysogenum DN16、Alternaria tenuissima DN09对G+细菌和G–大肠杆菌都有较好的抑制作用。其他真菌至少对6种指示细菌中的一种有抑制作用。未筛选到对水稻纹枯病真菌有抑菌活性的菌株。
| Isolates | Indicator | |||||
| B. subtilis | B. mycoides | S. aureus | A. baumanii | E. coli | P. aeruginosa | |
| Cladosporium cladosporioides DN01 (KY781386) | – | – | ++ | – | – | – |
| Cladosporium oxysporum DN02 (KY781385) | – | – | + | – | – | – |
| Cladosporium sphaerospermum DN03 (KY781384) | – | – | + | – | – | – |
| Plectosphaerella cucumerina DN04 (KY781383) | – | – | + | – | – | – |
| Alternaria tenuissima DN09 (KY781378) | + | – | – | – | ++ | – |
| Aspergillus versicolor DN11 (KY781376) | – | – | + | – | – | – |
| Alternaria longipes DN13 (KY781374) | – | + | – | – | – | – |
| Alternaria alternata DN14 (KY781373) | – | ++ | ++ | – | – | + |
| Stemphylium lycopersici DN15 (KY781372) | +++ | +++ | ++ | – | – | – |
| Penicillium chrysogenum DN16 (KY781371) | – | – | + | – | + | – |
| –: No transparent circle; +: 6≤Φ < 12 mm; ++: 12≤Φ < 18 mm; +++: Φ≥18 mm. Φ: Diameter of antimicrobe circle. The results were the average of three replicates. | ||||||
3 讨论
目前,对南海深海沉积物真菌的研究还比较少。本次研究分离到的真菌的ITS序列与数据库中已有真菌序列相似度都 > 97%。尽管采用了多种培养基和多种培养条件,但分离培养得到的真菌都属于子囊菌门和担子菌门,未分离到其他门的真菌。至今国内外还未有能通过培养的方式从深海沉积物中分离得到子囊菌门和担子菌门这两门以外的真菌,这可能与没有模拟深海环境(例如高压等)因素有关。可见,如何模拟深海环境分离得到新颖性高的深海真菌是一个急需解决的问题。分离培养到的属Cladosporium、Alternaria、Aspergillus、Penicillium被认为是陆源属真菌,但这些属的真菌也在多种深海环境中报道过,例如真菌Cladosporiumsp.在南海深海沉积物[3]、东印度洋深海沉积物[6, 23]、大西洋和太平洋深海热泉[5]都有发现。有报道称这个属的真菌(Cladosporium cladosporioides)能产不饱和脂肪酸来耐受低温[11]。真菌Aspergillus sp.在南海深海沉积物[2-3, 12]、印度洋深海沉积[32]、太平洋深海沉积物[4]都有发现,这个属的某些种所产的孢子能够在高压和低温的环境下存活[5],而且能参与到反硝化反应中[33]。Exophiala、Sarocladiu、Alternaria这3个属的真菌在多种深海环境中报道过[2-4, 32, 34]。Cystobasidium、Knufia这2个属的真菌在深海环境中被报道的较少,Singh等曾在印度洋深海沉积物中分离培养到这2个属的真菌[23]。实验中分离到的酵母Rhodotorula mucilaginosa普遍存在于多种深海环境,甚至在目前所知地球上最深的海沟——马里亚纳海沟中也发现了这株酵母的存在,并被多次报道用于抗冻蛋白的研究[6, 10, 34]。Stemphylium和Plectosphaerella两个属的真菌在深海环境中鲜少有报道,有报道称通过Illumina测序发现这个属分布于冲绳海沟的深海沉积物[34]。在本研究分离到的Cadophora这个属的真菌被报道存在于深海热泉,这个属的真菌同样被发现存在于南极环境,且是耐冷菌[5]。可见,本研究分离培养的所有真菌都曾在深海环境中被报道过。尽管一些真菌在陆地上也常有发现,但是通过适应深海环境可能具有了新的生理生化特性和功能,因此也具有极高的开发利用价值。为了排除分离培养过程中污染的可能性,所有实验步骤均在无菌操作台中进行,所有的器具和试剂均经过严格的灭菌,且实验时在操作台中设置了空白对照平板,这些平板上均无真菌生长;同时,本研究所有培养到的真菌属在其他深海环境中均有报道,这都说明本研究分离培养的真菌不是来自分离过程中的环境污染。
基于本研究的结果,4个站位的可培养真菌多样性存在较大差异。Zhang等[3]对南海9个站位的深海沉积物中可培养的真菌进行了比对,同样发现,各站位之间培养到的真菌多样性存在较大的差异。造成各站位之间真菌多样性差异的主要原因可能是各位点沉积物的营养组成不同[9];另一个可能的原因是,每个位点只有部分沉积物被用来分离培养真菌,这部分沉积物不能很好地代表这个站位[7]。对于每个站位的3个深度之间,本研究得到的结果是深度对真菌多样性没有显著的影响,这可能是3个深度之间跨度不大(才大约1 m)所致。Rédou等[24]研究了海底表层以下,跨度达千米的沉积物中可培养真菌的组成,发现深度对真菌多样性有显著的影响。因此,可培养真菌多样性和站位、深度之间的关系还有待进一步研究。
本实验中分离到的真菌展现出较好的抗菌活性,对至少一种细菌指示菌有抑制作用。除了已知能产多种化合物的Aspergillus和Penicillium属真菌外,本研究分离到的Cladosporium属(DN01、DN02、DN03) 真菌对金黄色葡萄球菌呈现良好抗性。Zhang等[3]同样从南海沉积物中分离到了这个属的真菌,其表现出抗真菌活性。我们分离培养到的Alternaria tenuissima DN09、Alternaria alternata DN14和Penicillium chrysogenum DN16不仅对G+细菌有良好的抗性,而且对G–的大肠杆菌和铜绿假单胞菌也表现出抑制作用。Zhang等[35]曾发现南海珊瑚来源的这个属对藤黄球菌有抑制作用。本研究中发现的有抗菌活性的真菌中,除Plectosphaerella sp.和Stemphylium sp.以外都曾被报道从南海沉积物中被分离培养,且有产活性物质的潜能[3, 13-22]。由此可见,南海沉积物来源的真菌是用于挖掘天然活性产物的良好资源。尤其值得一提的是,Stemphylium lycopersici DN15对B. subtilis、B. mycoides和S. aureus具有较强的抑制活性,非常值得进一步对其次级代谢产物进行研究。Teiten等[36]从陆地植物来源的一株匐柄霉属(Stemphylium)真菌中分离出Altersolanol A,这种物质有抗肿瘤活性。Zhou等[37]从南海红树林来源的这个属的真菌中分离出吡喃酮派生物,这种物质有抗多种细菌的活性。总之,该研究采用多种分离方法与策略,揭示了南海多个站位、不同深度的深海沉积物中可培养真菌的多样性。同时,得到了多株具有抗菌活性的真菌,尤其是具有多种抗菌活性的Stemphylium lycopersici DN15、Alternaria tenuissima DN09、Alternaria alternata DN14、Penicillium chrysogenum DN16。这些结果不仅丰富了我们对深海可培养真菌多样性的了解,同时也为尽快一步开发利用深海真菌资源提供了依据。
致谢:感谢上海交通大学王风平教授提供深海沉积物样品,何亚文教授提供水稻纹枯病真菌(Rhizotonia solani)。
| [1] | Roth Jr FJ, Orpurt PA, Ahearn DG. Occurrence and distribution of fungi in a subtropical marine environment. Canadian Journal of Botany, 1964, 42(4): 375-383. DOI:10.1139/b64-037 |
| [2] | Lai XT, Cao LX, Tan HG, Fang S, Huang YL, Zhou SN. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. The ISME Journal, 2007, 1(8): 756-762. DOI:10.1038/ismej.2007.51 |
| [3] | Zhang XY, Zhang Y, Xu XY, Qi SH. Diverse deep-sea fungi from the South China Sea and their antimicrobial activity. Current Microbiology, 2013, 67(5): 525-530. DOI:10.1007/s00284-013-0394-6 |
| [4] | Xu W, Pang KL, Luo ZH. High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microbial Ecology, 2014, 68(4): 688-698. DOI:10.1007/s00248-014-0448-8 |
| [5] | Burgaud G, Le Calvez T, Arzur D, Vandenkoornhuyse P, Barbier G. Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environmental Microbiology, 2009, 11(6): 1588-1600. DOI:10.1111/emi.2009.11.issue-6 |
| [6] | Zhang XY, Tang GL, Xu XY, Nong XH, Qi SH. Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS One, 2014, 9(10): e109118. DOI:10.1371/journal.pone.0109118 |
| [7] | Singh P, Raghukumar C, Meena RM, Verma P, Shouche Y. Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecology, 2012, 5(5): 543-553. DOI:10.1016/j.funeco.2012.01.001 |
| [8] | Damare S, Raghukumar C, Raghukumar S. Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Research Part Ⅰ:Oceanographic Research Papers, 2006, 53(1): 14-27. DOI:10.1016/j.dsr.2005.09.005 |
| [9] | Singh P, Raghukumar C, Verma P, Shouche Y. Fungal community analysis in the deep-sea sediments of the Central Indian Basin by culture-independent approach. Microbial Ecology, 2011, 61(3): 507-517. DOI:10.1007/s00248-010-9765-8 |
| [10] | Singh P, Raghukumar C, Verma P, Shouche Y. Assessment of fungal diversity in deep-sea sediments by multiple primer approach. World Journal of Microbiology and Biotechnology, 2012, 28(2): 659-667. DOI:10.1007/s11274-011-0859-3 |
| [11] | Zhang T, Wang NF, Zhang YQ, Liu HY, Yu LY. Diversity and distribution of fungal communities in the marine sediments of Kongsfjorden, Svalbard (High Arctic). Scientific Reports, 2015, 5: 14524. DOI:10.1038/srep14524 |
| [12] |
Qu J, Liu KH, Ding XW, Deng BW, Chen WQ, Guo QL, Tian XP, Zhang S, Li WJ. Fungal diversity and enzyme activities in marine sediments in the South China Sea. Acta Microbiologica Sinica, 2014, 54(5): 552-562.
(in Chinese) 曲佳, 刘开辉, 丁小维, 邓百万, 陈文强, 郭庆兰, 田新朋, 张偲, 李文均. 南海局部海洋沉积物中真菌多样性及产酶活性. 微生物学报, 2014, 54(5): 552-562. |
| [13] | Li CS, Li XM, Gao SS, Lu YH, Wang BG. Cytotoxic anthranilic acid derivatives from deep sea sediment-derived fungus Penicillium paneum SD-44. Marine Drugs, 2013, 11(8): 3068-3076. DOI:10.3390/md11083068 |
| [14] | Wang MH, Li XM, Li CS, Ji NY, Wang BG. Secondary metabolites from Penicillium pinophilum SD-272, a marine sediment-derived fungus. Marine Drugs, 2013, 11(6): 2230-2238. DOI:10.3390/md11062230 |
| [15] | Shang Z, Li XM, Meng L, Li CS, Gao SS, Huang CG, Wang BG. Chemical profile of the secondary metabolites produced by a deep-sea sediment-derived fungus Penicillium commune SD-118. Chinese Journal of Oceanology and Limnology, 2012, 30(2): 305-314. DOI:10.1007/s00343-012-1075-1 |
| [16] | You JL, Dai HQ, Chen ZH, Liu GJ, He ZX, Song FH, Yang X, Fu HA, Zhang LX, Chen XP. Trichoderone, a novel cytotoxic cyclopentenone and cholesta-7, 22-diene-3β, 5α, 6β-triol, with new activities from the marine-derived fungus Trichoderma sp.. Journal of Industrial Microbiology & Biotechnology, 2010, 37(3): 245-252. |
| [17] | Lin XP, Zhou XF, Wang FZ, Liu KS, Yang B, Yang XW, Peng Y, Liu J, Ren Z, Liu YH. A new cytotoxic sesquiterpene quinone produced by Penicillium sp. F00120 isolated from a deep sea sediment sample. Marine Drugs, 2012, 10(1): 106-115. |
| [18] | Li CS, An CY, Li XM, Gao SS, Cui CM, Sun HF, Wang BG. Triazole and dihydroimidazole alkaloids from the marine sediment-derived fungus Penicillium paneum SD-44. Journal of Natural Products, 2011, 74(5): 1331-1334. DOI:10.1021/np200037z |
| [19] | Peng J, Zhang XY, Tu ZC, Xu XY, Qi SH. Alkaloids from the deep-sea-derived fungus Aspergillus westerdijkiae DFFSCS013. Journal of Natural Products, 2013, 76(5): 983-987. DOI:10.1021/np400132m |
| [20] | Wang FZ, Huang Z, Shi XF, Chen YC, Zhang WM, Tian XP, Li J, Zhang S. Cytotoxic indole diketopiperazines from the deep sea-derived fungus Acrostalagmus luteoalbus SCSIO F457. Bioorganic & Medicinal Chemistry Letters, 2012, 22(23): 7265-7267. |
| [21] | Chen ZM, Huang HB, Chen YC, Wang ZW, Ma JY, Wang B, Zhang WM, Zhang CS, Ju JH. New cytochalasins from the marine-derived fungus Xylaria sp. SCSIO 156. Helvetica Chimica Acta, 2011, 94(9): 1671-1676. DOI:10.1002/hlca.v94.9 |
| [22] | Fredimoses M, Zhou XF, Lin XP, Tian XP, Ai W, Wang JF, Liao SR, Liu J, Yang B, Yang XW, Liu YH. New prenylxanthones from the deep-sea derived fungus Emericella sp. SCSIO 05240. Marine Drugs, 2014, 12(6): 3190-3202. DOI:10.3390/md12063190 |
| [23] | Singh P, Raghukumar C, Verma P, Shouche Y. Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Diversity, 2010, 40(1): 89-102. DOI:10.1007/s13225-009-0009-5 |
| [24] | Rédou V, Navarri M, Meslet-Cladière L, Barbier G, Burgaud G. Species richness and adaptation of marine fungi from deep-subseafloor sediments. Applied and Environmental Microbiology, 2015, 81(10): 3571-3583. DOI:10.1128/AEM.04064-14 |
| [25] | Jebaraj CS, Raghukumar C, Behnke A, Stoeck T. Fungal diversity in oxygen-depleted regions of the Arabian Sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiology Ecology, 2010, 71(3): 399-412. DOI:10.1111/fem.2010.71.issue-3 |
| [26] | Nagahama T, Nagano Y. Cultured and uncultured fungal diversity in deep-sea environments//Raghukumar C. Biology of Marine Fungi. Berlin Heidelberg:Springer, 2012:173-187. |
| [27] | Tsuji M, Fujiu S, Xiao N, Hanada Y, Kudoh S, Kondo H, Tsuda S, Hoshino T. Cold adaptation of fungi obtained from soil and lake sediment in the Skarvsnes ice-free area, Antarctica. FEMS Microbiology Letters, 2013, 346(2): 121-130. DOI:10.1111/fml.2013.346.issue-2 |
| [28] | Collado J, Platas G, Paulus B, Bills GF. High-throughput culturing of fungi from plant litter by a dilution-to-extinction technique. FEMS Microbiology Ecology, 2007, 60(3): 521-533. DOI:10.1111/fem.2007.60.issue-3 |
| [29] |
Zhang D, Jiang Q, Li YX, Feng GF, Sun W, Zhang FL, Li ZY. Diversity and antibacterial activity of fungi associated with South China Sea sponges and corals. Chinese Journal of Marine Drugs, 2015, 34(2): 1-9.
(in Chinese) 张丹, 蒋群, 李英新, 冯国芳, 孙伟, 张风丽, 李志勇. 中国南海海绵和珊瑚共附生真菌多样性及其抑菌活性研究. 中国海洋药物, 2015, 34(2): 1-9. |
| [30] | Singh R, Jain A, Panwar S, Gupta D, Khare SK. Antimicrobial activity of some natural dyes. Dyes and Pigments, 2005, 66(2): 99-102. DOI:10.1016/j.dyepig.2004.09.005 |
| [31] |
Su XY, Li J, Shi GL, Wu J, Wen ZY, Wang YN. Antifungal activities of the extracts from walnut peel against six phytopathogenic fungi. Journal of Beijing University of Agriculture, 2008, 23(1): 42-44.
(in Chinese) 苏学友, 李疆, 师光禄, 吴静, 温哲屹, 王有年. 核桃青皮提取物对6种植物病原真菌的抑菌活性研究. 北京农学院学报, 2008, 23(1): 42-44. |
| [32] | Raghukumar C, Raghukumar S, Sheelu G, Gupta SM, Nath BN, Rao BR. Buried in time:culturable fungi in a deep-sea sediment core from the Chagos Trench, Indian Ocean. Deep Sea Research Part Ⅰ:Oceanographic Research Papers, 2004, 51(11): 1759-1768. DOI:10.1016/j.dsr.2004.08.002 |
| [33] | Cathrine SJ, Raghukumar C. Anaerobic denitrification in fungi from the coastal marine sediments off Goa, India. Mycological Research, 2009, 113(1): 100-109. DOI:10.1016/j.mycres.2008.08.009 |
| [34] | Zhang XY, Wang GH, Xu XY, Nong XH, Wang J, Amin M, Qi SH. Exploring fungal diversity in deep-sea sediments from Okinawa Trough using high-throughput Illumina sequencing. Deep Sea Research Part Ⅰ:Oceanographic Research Papers, 2016, 116: 99-105. DOI:10.1016/j.dsr.2016.08.004 |
| [35] | Zhang XY, Bao J, Wang GH, He F, Xu XY, Qi SH. Diversity and antimicrobial activity of culturable fungi isolated from six species of the South China Sea gorgonians. Microbial Ecology, 2012, 64(3): 617-627. DOI:10.1007/s00248-012-0050-x |
| [36] | Teiten MH, Mack F, Debbab A, Aly AH, Dicato M, Proksch P, Diederich M. Anticancer effect of altersolanol A, a metabolite produced by the endophytic fungus Stemphylium globuliferum, mediated by its pro-apoptotic and anti-invasive potential via the inhibition of NF-κB activity. Bioorganic & Medicinal Chemistry, 2013, 21(13): 3850-3858. |
| [37] | Zhou XM, Zheng CJ, Song XP, Han CR, Chen WH, Chen GY. Antibacterial α-pyrone derivatives from a mangrove-derived fungus Stemphylium sp. 33231 from the South China Sea. The Journal of Antibiotics, 2014, 67(5): 401-403. DOI:10.1038/ja.2014.6 |
 2017, Vol. 57
2017, Vol. 57