中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 郑文韬, 张友明, 卞小莹. 2017
- Wentao Zheng, Youming Zhang, Xiaoying Bian. 2017
- Red/ET同源重组技术及其在微生物基因组挖掘中的应用进展
- Advances in Red/ET recombineering and its application for microbial genome mining
- 微生物学报, 57(11): 1735-1746
- Acta Microbiologica Sinica, 57(11): 1735-1746
-
文章历史
- 收稿日期:2017-07-04
- 修回日期:2017-08-12
- 网络出版日期:2017-08-22
Xiaoying Bian, Tel:+86-532-67721928, E-mail:bianxiaoying@sdu.edu.cn
1998年,Zhang等发现大肠杆菌(Escherichia coli) Rac原噬菌体上的操纵子recE/recT编码的蛋白能够高效地介导体内同源重组的发生[1],而且Muyers等发现来源于大肠杆菌λ噬菌体的Redα/Redβ蛋白与RecE/RecT蛋白有类似的介导同源重组的功能[2],Stewart等将Redα/Redβ和RecE/RecT重组酶体系合并命名为“Red/ET重组工程(Red/ET recombineering)”[3]。与体内自然发生的同源重组相比,Red/ET重组系统只需要40–50 bp的同源臂就能实现高效的同源重组,这一点突破了酶切位点和长片段PCR扩增的局限性对传统基因操作技术的限制,通过合成引物的方式将短同源臂加在PCR产物的两端,可以与任意位置的目标序列进行同源重组,同时Red/ET同源重组技术的操作过程在大肠杆菌中完成,极大缩短了构建载体和基因修饰的步骤与时间,因此Red/ET同源重组技术的应用已经越来越广泛[4-5]。我们简要综述了2010年以来Red/ET同源重组技术在大肠杆菌及其他细菌中的研究进展,而且对其用于微生物基因组挖掘,尤其是微生物基因簇的异源表达领域的应用进展做概况介绍。
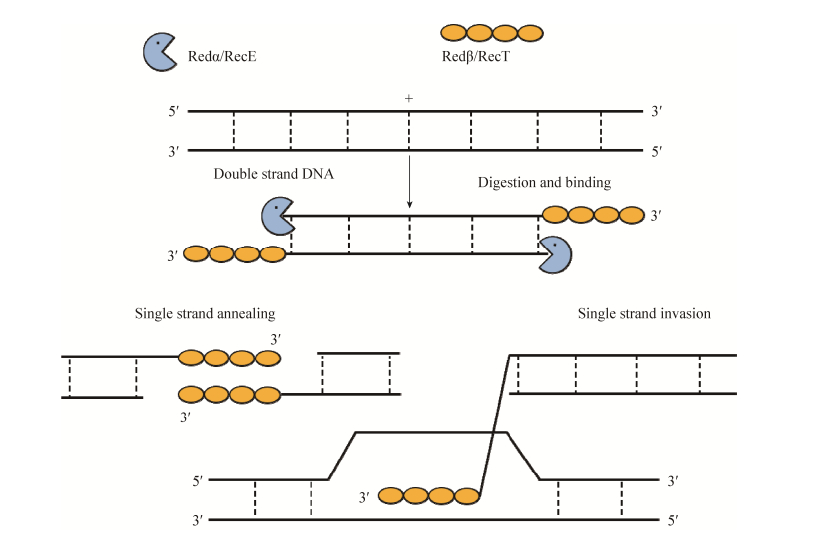
1 大肠杆菌中Red/ET同源重组技术的研究进展Red/ET重组系统工作时,首先由具有5′→3′端的核酸外切酶活性的RecE和Redα从5′端开始酶切双链DNA (dsDNA)分子[6],产生局部的3′端单链DNA (ssDNA,single-stranded DNA)并马上与具有单链结合蛋白活性的RecT或Redβ结合,防止单链被降解并在RecT或Redβ帮助下寻找到同源序列[7-8],完成重组(图 1)。来源于λ噬菌体的Redαβ蛋白对能够高效地介导线性DNA片段与环状DNA分子之间的线环重组(linear plus circular homologous recombination, LCHR,图 2-A),而全长的RecE和RecT蛋白对更为有效地介导2个线性DNA片段之间的重组(线线重组,linear plus linear homologous recombination, LLHR,图 2-B),对于2个2 kb的小片段线线同源重组(图 2),全长的RecET比RecE602T (RecE602是全长的RecE蛋白删减上游与Redα不同源的602个氨基酸)的重组效率高322倍,比Redαβ高36倍[9]。Redαβ介导的线环重组是发生在复制叉,需要DNA复制的进程,而RecET介导的线线重组不依赖复制进程,说明线线重组的机理跟线环重组是不同的。而且利用RecET介导的线线重组技术不仅能够从细菌人工染色体(BAC)上精准地亚克隆基因片段,而且能够从酶切后的原核生物基因组DNA上高效地一步直接克隆出大片段目标DNA序列(比如次级代谢产物生物合成基因簇等),不需要费时费力的基因文库构建和筛选,极大地促进了后基因组时代的大型基因的克隆研究,意义重大,受到众多科学家的关注[9-11]。
由λ Red噬菌体的单链结合蛋白Redβ介导的多元自动基因工程(multiplex automated genome engineering,MAGE)通过程序化循环,同时对染色体上多个位置进行单链重组,从而产生多种基因组组合。2009年Wang等利用MAGE技术使用混合的单链DNA同时对大肠杆菌中1-脱氧-D-木酮糖-5-磷酸酯(DXP)的生物合成途径的24个位点进行遗传修饰,平均每天可产生43亿个特定位点的基因突变型组合,通过高通量筛选从中获得了能够用来进行大量生产具有工业用途的类异戊二烯番茄红素的突变株[12]。2011年Farren使用MAGE技术在大肠杆菌中进行同义密码子的替换,将314个TAG终止密码子同时替换为同义的TAA密码子,从而推测出大肠杆菌个体的重组频率并对有关联的表型进行确定[13]。之后在2012年Wang等将MAGE与抗性筛选结合的“共选择”MAGE技术(Cos-MAGE)应用到芳香氨基酸衍生物的生物合成优化中,将T7启动子同时插入到12个基因组的操纵子中,以此为基础可以对内源性的异位显性进行探测并改造遗传途径[14]。
Red/ET重组工程结合反向筛选技术,可以对DNA进行无痕敲除、定点突变和模块替换等修饰。细菌人工染色体(BAC)在研究基因和蛋白功能上有明显优势,但是无痕修饰与精确点突变较难,2011年Bird等使用rpsL-neo反向筛选组件在进行BACs修饰时,发现重复序列之间的分子内重组是造成修饰失败的主要原因,他们推测是λ Red系统促进了分子内重组的产生,在第二步重组时单独表达Redβ,大部分rpsL-neo反向筛选组件可以被单链核苷酸链替换,利用这个方法,他们在敲除内源性TACC3基因后,通过将修饰后的TACC3基因转入人体细胞来鉴定磷酸化特异性纺锤体缺陷的问题[15]。利用大肠杆菌毒素基因ccdB及其解毒剂基因ccdA的相互作用可以对大肠杆菌的基因组或者质粒进行精确的无痕修饰,其中ccdB作为反向选择标记。2014年Wang等将ccdB-amp组件插入到大肠杆菌的高拷贝质粒上,而将ccdA解毒基因与Red操纵子构建在温敏型质粒上,通过两步重组实现了含有重复序列的多拷贝质粒上的非核糖体多肽合成酶基因簇plu3263的定点突变,成功修饰后的突变菌株中不再产生luminmide[16]。
Red/ET同源重组系统是细菌中基因操作的有效工具,但很多因素限制了重组的效率,2012年Joshua等鉴定了大肠杆菌内源性核酸酶ExoVII可以降解单链或双链DNA的末端,移除这个核酸酶可以提高重组效率,而将5个核酸外切酶(RecJ、ExoI、ExoVII、ExoX和λExo)敲除可以提高Cos-MAGE的效率[17]。噬菌体重组酶介导的同源重组系统能够进行高效的染色体基因修饰,两端带有短同源臂的单链或双链DNA在重组酶介导下可以进行精确的基因修饰。2016年Lynn等的研究表明Red/ET重组系统在介导重组时需要暴露同源互补单链,这种单链区域可能在DNA复制时或者双链DNA经单链核酸酶消化后产生[18]。单链退火蛋白(SSAPs)如Redβ通过促进互补DNA链的退火介导同源重组,同一年Sivaraman等发现删除Redβ的C端仍能促进退火和核蛋白丝的形成,但不能介导同源重组,免疫共沉淀实验证明双链DNA的重组涉及Redα-Redβ的蛋白相互作用,该相互作用需要Redβ的C端区域和N端区域的共同参与[19]。
2 Red/ET同源重组技术在其他细菌中的应用以噬菌体蛋白为基础构建的同源重组系统在细菌基因修饰中应用广泛,不仅可以在大肠杆菌中高效地修饰DNA序列,同样可以在其他多种细菌中使用(表 1)。2003年Hu等将大肠杆菌λ Red重组系统以复制质粒形式导入痢疾杆菌(Shigella flexneri)中,通过表达重组酶用带有40–50 bp同源臂的抗性基因替换其染色体上的alkA基因[20]。2005年Karlinsey等利用大肠杆菌λ Red重组系统对鼠伤寒沙门氏菌(Salmonella enterica)进行基因修饰,将带有40 bp同源臂的tetRA基因(携带四环素抗性)替换其染色体的fliA基因和flgM基因,再将风产液菌(Aquifex aeolicus)和沙眼衣原体(Chlamydia trachomatis)的同源基因替换插入的tetRA基因,为从遗传学角度研究物种间蛋白关系奠定基础[21]。2013年Hu等在根癌农杆菌(Agrobacterium tumefaciens)中利用大肠杆菌λ Red重组系统使用带有70 bp同源臂的卡那霉素抗性基因成功替换单拷贝质粒Ti上的virG基因和染色体上的crdS基因[22]。噬菌体介导的同源重组系统在基因组替换和敲除上效果显著,Kang等的研究表明在能够自然转化的伯克氏菌中,大肠杆菌λ Red重组系统可以进行基因敲除、插入和亚克隆,他们利用λ Red重组系统分别从泰国伯克氏菌(B. thailandensis)和类鼻疽伯克氏菌(B. pseudomallei)中通过亚克隆得到了代谢阿拉伯糖的操纵子(11.6 kb)和malleobactin的生物合成基因簇(31.5 kb)[23]。
| Bacteria | Proteins | Application | Reference |
| Shigella flexneri | E. coli λ Red | Delection of alkA | [20] |
| Salmonella enterica | E. coli λ Red | Replacement of fliA, flgM | [21] |
| Agrobacterium tumefaciens | E. coli λ Red | Replacement of virG, crdS | [22] |
| Burkhodleria thailandensis B. pseudomallei | E. coli λ Red | Cloning of the arabinose utilization operon and siderophore malleobactin biosynthetic cluster | [23] |
| Mycobacterium smegmatis | gp60/61 in host phage Che9c | The inactivation of the hygromycin gene on the plasmid | [24-25] |
| Lactobacillus reuteri | Native recT1、recT2 | Mutations were mediated by single strand recombination | [26] |
| Pseudomonas syringae | RecETpsy in Pseudomonas syringae pv. syringae B728a | 500 ng substrate is sufficient to mediate homologous recombination | [27] |
| Photorhabdus luminescens | Plu2935-Plu2936 in host phage | The recombination efficiency was nearly 5 times higher than that using Redα/β/γ | [28] |
来自大肠杆菌噬菌体的Red/ET重组系统只能在部分与大肠杆菌亲缘关系较近的革兰氏阴性菌中应用,而在与其亲缘关系较远的菌类中,该系统的功能是受限制的。然而Red/ET重组系统的建立却为我们在其他细菌基因组或者其噬菌体基因组中寻找类似的重组酶蛋白提供了理论依据和技术指导,目前已有多种利用细菌自身噬菌体或原噬菌体的重组蛋白建立起的具有种属特异性同源重组工程系统。2007年Kessel等采用来自革兰氏阳性细菌耻垢分枝杆菌(Mycobacterium smegmatis)噬菌体Che9c中的Redα/β类似蛋白gp60/61,构建了分枝杆菌特异性的重组工程系统,能够以带有50 bp短同源臂的单链寡核苷酸作为底物进行同源重组,通过点突变失活质粒上的潮霉素抗性基因,在耻垢分枝杆菌和结核分枝杆菌中的重组效率分别可达到104 CFU/μg DNA和103 CFU/μg DNA,而来源于大肠杆菌的RecET和Redαβγ则不能在分枝杆菌中有效介导单链重组[24-25]。2012年Pijkeren等在罗伊氏乳杆菌(Lactobacillus reuteri)中找到与recT同源的2个基因recT1和recT2,2个基因编码的蛋白能够在乳酸杆菌中有效地介导单链重组的发生[26]。2010年Bryan等从丁香假单胞菌的噬菌体或者原噬菌体残留物(Pseudomonas syringae pv. syringae B728a)中发现了与大肠杆菌RecE和RecT同源的一种新的重组-核酸外切酶组合(RecETpsy),该组合能够促进染色体中特定位点与线性DNA底物之间的高效同源重组,对RecETpsy介导的同源重组条件进行优化时发现500 ng底物足够诱发同源重组,而将同源臂由100 bp增加至500 bp时重组效率提高了250倍[27]。2015年Yin等采用发光杆菌(Photorhabdus luminescens)原噬菌体中的Redα/β类似蛋白Plu2935/Plu2936构建了发光杆菌和嗜线虫致病杆菌(Xenorhabdus stockiae)的重组工程系统,并且首次发现了Plu2935上游的Plu2934的功能跟Redγ类似,在明亮发光杆菌中利用Plu2934-35-36重组工程系统对基因组进行修饰,重组效率比采用Redα/β/γ提高了将近5倍[28]。
3 Red/ET同源重组技术在微生物基因组挖掘中的应用微生物次级代谢产物是最重要的药物和生物农药来源之一,从生物合成的角度来说,编码同一次级代谢产物合成酶的基因通常都集中在基因组的一个区域,所以又称为生物合成基因簇,这些基因簇一般在10–120 kb左右,而且已经在许多细菌中发现。随着基因组测序技术的发展,生物信息学分析发现微生物所蕴藏的天然产物资源被大大低估,细菌所具有的基因簇数量要远远大于目前已知的天然产物的数量[29-30],微生物中还有大量的“暗物质(未知基因簇)”有待发掘,仍然具有产生新颖先导结构的巨大潜力和空间[31-32]。基因组挖掘是以基因簇序列和生物合成途径为导向发现天然产物的策略,能够发现更多新颖化合物用于进行先导药物的筛选[33]。现今基因组挖掘的策略按照宿主的不同分为在本源菌中的同源表达和在异源宿主中的异源表达[34]。前者主要依赖在本源菌中的遗传操作激活生物合成基因簇,通过代谢谱比较发现新的化合物。但是由于大多数本源菌都难以遗传操作,这种策略有时显得费时费力,因此需在本源菌中构建流畅的遗传操作体系,便于微生物基因资源的深度开发。Yin等在发光杆菌和嗜线虫致病杆菌构建了基于自身噬菌体的Red/ET重组工程系统,并利用该技术快速准确地插入四环素诱导型启动子,激活了49 kb的非核糖体多肽合成酶基因簇Plu2670[28, 35],为进一步开发发光杆菌和嗜线虫杆菌的天然产物奠定了技术基础。
微生物次生代谢产物生物合成途径在基因组上的成簇分布为异源表达策略提供了可能。该策略需要将基因簇完整地克隆到穿梭载体上然后稳定地转移到合适简单的异源宿主中功能表达[36]。大多数微生物次级代谢产物的生物合成基因簇比较大,黏粒等基因文库的构建通常不能包括完整的基因簇,需要利用Red/ET同源重组技术将来源于不同黏粒的基因簇片段缝合到一个载体上,然后插入转移基因、启动子等元件,用于基因簇在不同宿主间的转移和在异源宿主中的功能性表达(图 3-A,表 2)。Wenzel等利用Red/ET同源重组技术成功缝合两个黏粒,从而获得了完整的myxochromide S合成基因簇(43 kb),并且在恶臭假单胞菌中实现了异源表达[37]。同样地,Perlova等使用Red/ET同源重组技术及基因缝合技术获得了来自橙色标桩菌(Stigmatella aurantiaca) DW4-3/1完整的myxothiazol合成基因簇(57 kb)并在黄色黏球菌(Myxococcus xanthus)中实现了异源表达[38];Binz等将链霉菌Streptomyces sp. Tü6071的phenalinolactone合成基因簇(42 kb)从2个质粒中亚克隆下来并通过基因缝合技术获得完整基因簇,在多个链霉菌中实现了异源表达[39];Hu等将链霉菌(S. refuineus)的anthramycin基因簇(32.5 kb)利用Red/ET同源重组技术从2个质粒中缝合到一起,在变铅青链霉菌中实现了异源表达[40];Wolpert等将链霉菌(S. rishiriensis) DSM40489的coumermycin A1基因簇(36.8 kb)缝合并在天蓝色链霉菌中异源表达[41];Fu等将纤维堆囊菌(Sorangium cellulosum)的埃博霉素基因簇(58 kb)从2个质粒中缝合到一起并在黄色黏球菌中异源表达[42],同样还在新的异源表达宿主伯克氏菌DSM7029中成功表达[43];Chai等将孢囊杆菌(Cystobacter sp.)SBCb004的tubulysin基因簇(40 kb)从2个质粒中缝合到一起并在黄色黏球菌和恶臭假单胞菌中异源表达[44];Kolinko等将磁螺菌(Magnetospirillum gryphiswaldense)中合成磁小体的基因簇片段通过表达Red/ET重组系统进行缝合、修饰等,并在深红红螺菌(Rhodospirillum rubrum)中异源表达磁小体[45]。
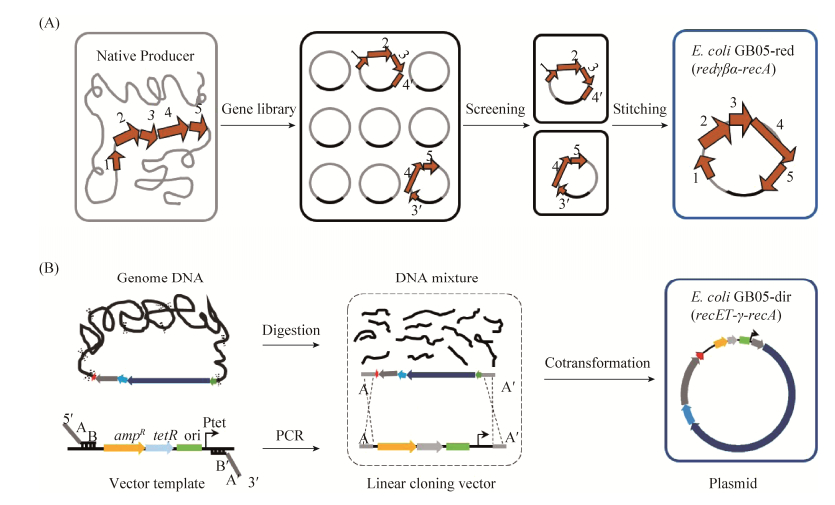
|
| 图 3 Red/ET同源重组技术介导的大型生物合成基因簇的克隆[37, 50] Figure 3 Red/ET recombineering mediated cloning of large biosynthetic gene clusters[37, 50]. A: Gene stitching based on gene library and Red/ET recombineering; B: Red/ET recombineering-mediated LLHR for direct cloning of large gene clusters from genomic DNA without construction of gene library. |
| Compound | Native host | Gene cluster size/kb | Method | Heterologous host | Reference |
| Myxochromide S | Stigmatella aurantiaca | 43.0 | Gene library and stitching | Pseudomonas putida Myxococcus xanthus | [37, 57] |
| Myxothiazol | Stigmatella aurantiaca DW4-3/1 | 57.0 | Gene library and stitching | Myxococcus xanthus | [38] |
| Phenalinolactone | Streptomyces sp. Tü6071 | 42.0 | Gene library and stitching | Streptomyces | [39] |
| Coumermycin A1 | Streptomyces rishiriensis DSM40489 | 36.8 | Gene library and stitching | Streptomyces coelicolor M512 | [41] |
| Epothilone | Sorangium cellulosum | 58.0 | Gene library and stitching | Myxococcus xanthus, Burkholderia DSM7029 | [42-43] |
| Tubulysin | Cystobacter sp. SBCb004 | 40.0 | Gene library and stitching | Myxococcus xanthus, Pseudomonas putida | [44] |
| Magnetosome | Magnetospirillum gryphiswaldense | 26.0 | Gene library and direct cloning | Rhodospirillum rubrum | [45] |
| Disorazol A | Sorangium cellulosum | 58.0 | Gene library and modification | Myxococcus xanthus | [49] |
| Luminmide | Photorhabdus luminescens TT01 | 15.6 | LLHR direct cloning | E. coli GB05-MtaA | [47] |
| Luminmycin | Photorhabdus luminescens TT01 | 18.3 | LLHR direct cloning | E. coli Nissle 1917 | [51] |
| Syringolin | Pseudomonas syringae pv. syringae B301D-R | 19.0 | LLHR direct cloning | E. coli GB05-MtaA | [50] |
| Glidobactin | Burkholderia DSM7029 | 28.0 | LLHR direct cloning | E. coli Nissle 1917 | [50] |
| Salinomycin | Streptomyces albus DSM41398 | 106.0 | LLHR direct cloning and stitching | Streptomyces coelicolor | [53] |
| Bacillomycin | Bacillus amyloliquefaciens FZB42 | 37.2 | LLHR direct cloning | Bacillus subtilis | [52] |
| Edeine | Brevibacillus brevis X23 | 48.3 | LLHR direct cloning | [52] |
利用RecET同源重组技术对基因簇进行修饰是提高天然产物产量的方法之一,尤其是启动子更换能很大程度上提高化合物的产量[46]。Ongley等利用Red/ET技术在不可纯培养的海洋藻青菌(Moorea producens)中lyngbyatoxin的合成基因簇中加入启动子和核糖体结合位点,使之在亲缘关系较远的大肠杆菌GB05-MtaA菌株中成功表达产生lyngbyatoxin A和其前体物indolactam-V,未经优化的产量可以达到25.6 mg/L和150.0 mg/L[47]。Tu等将纤维堆囊菌的disorazol A基因簇(58 kb)从两个质粒中缝合到一起,通过Ptet启动子控制合成基因簇的表达,在disD基因上游插入人工合成的启动子Pcp25[48],使其在黄色黏球菌中的产量提高了7倍[49]。
Red/ET同源重组技术还能免建库从基因组中直接克隆大型基因簇(图 3-B,表 2),大大促进了微生物次级代谢产物的异源表达研究。2012年,Fu等报道了全长RecE和RecT能够在大肠杆菌中免建库直接克隆大片段基因簇,利用这一技术,从发光杆菌中直接克隆得到10个未知基因簇(10–50 kb),其中plu3263 (15.6 kb)和plu1881-plu1877 (18.3 kb) 2个基因簇分别在大肠杆菌菌株中实现了异源表达并获得luminmides和luminmycin两种化合物[9, 51],其中luminmycin基因簇在原始菌中是沉默的,我们通过异源表达和启动子更换成功激活了这个沉默基因簇[51];我们还利用该方法从丁香假单胞菌和伯克氏菌DSM7029中直接克隆到syringolin (19 kb)和glidobactin (28 kb)的合成基因簇,并分别在大肠杆菌GB05-MtaA菌株和Nissle 1917中实现了异源表达并发现了多个新的化合物[46, 50]。Liu等利用Red/ET同源重组技术从解淀粉芽孢杆菌(Bacillus amyloliquefaciens) FZB42中克隆到了bacillomycin合成基因簇(37.2 kb),然后进一步利用该技术将克隆到的基因簇整合到枯草芽孢杆菌的基因组上并成功实现了bacillomycin的异源表达,同时他们还从短短芽孢杆菌(Brevibacillus brevis) X23中成功直接克隆到了抗菌物质edeine的生物合成基因簇(48.3 kb)[52]。对于较大的生物合成基因簇片段( > 50 kb),需要多次直接克隆结合基因缝合技术。2015年Yin等先利用直接克隆技术分3次从白色链霉菌(S. albus) DSM41398中得到盐霉素基因簇(106 kb)的3个片段(F1,43 kb;F2,33 kb;F3,30 kb),然后利用基因缝合技术对3个片段进行缝合和组装,得到了完整的盐霉素合成基因簇并在天蓝色链霉菌中实现了异源表达[53]。最近,我们发布了免建库直接克隆技术的详细方法及标准化克隆载体、转移基因盒和多功能启动子等,可以实现克隆载体、转移基因和启动子快速切换,大大加快了直接克隆技术在微生物基因组挖掘中的应用[54]。
Red/ET同源重组技术不仅在挖掘未知化合物方面能够发挥重要的作用,在研究化合物合成机理上也有突出的表现。带有colibactin合成基因簇的细菌感染真核细胞会引起后者体内双链DNA的断裂,但是用菌体上清液或者是溶菌物对真核细胞进行作用则不会引发该现象,Bian等利用Red/ET技术对colibactin的合成后修饰过程进行阻断,确定了colibactin的前体物质[55]。与之类似,Bian等还利用Red/ET同源重组技术结合ccdB反向筛选标记对luminmide合成基因簇的腺苷酰化结构域进行点突变,通过检测化合物产量的变化来确定这些点突变能够改变次级代谢产物的组分,有利于后期的分离纯化工作[56]。
4 总结Red/ET同源重组技术不仅可以对大肠杆菌中DNA分子的任意位置进行精确的修饰,包括单碱基改变和基因的插入与删除,而且现在已经能够在多个其他细菌中工作。将来继续发现新的宿主特异性重组酶,并使之能够在更多的细菌中(假单胞菌、芽孢杆菌、伯克氏菌等)建立以Red/ET同源重组技术为基础的高效遗传操作体系,促进微生物功能基因组和基因组挖掘的发展。以全长RecET介导的线线重组为基础的免建库直接克隆技术为大片段基因簇的研究奠定了基础,极大促进了后基因组时代以异源表达为策略的微生物次级代谢产物的发现和开发。
随着分子生物学和基因工程技术的发展,Red/ET同源重组技术也在不断发展和完善,已经成为基因功能研究中一项热门的技术,在基因组学研究和微生物基因组功能挖掘中将扮演更重要的角色。
| [1] | Zhang YM, Buchholz F, Muyrers JPP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genetics, 1998, 20(2): 123-128. DOI:10.1038/2417 |
| [2] | Muyrers JPP, Zhang YM, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Research, 1999, 27(6): 1555-1557. DOI:10.1093/nar/27.6.1555 |
| [3] | Zhang YM, Muyrers JPP, Rientjes J, Stewart AF. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Molecular Biology, 2003, 4(1): 1. DOI:10.1186/1471-2199-4-1 |
| [4] | Muyrers JPP, Zhang YM, Stewart AF. Techniques:recombinogenic engineering-new options for cloning and manipulating DNA. Trends in Biochemical Sciences, 2001, 26(5): 325-331. DOI:10.1016/S0968-0004(00)01757-6 |
| [5] | Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering:a homologous recombination-based method of genetic engineering. Nature Protocols, 2009, 4(2): 206-223. DOI:10.1038/nprot.2008.227 |
| [6] | Little JW. An exonuclease induced by bacteriophage λ Ⅱ. Nature of the enzymatic reaction. Journal of Biological Chemistry, 1967, 242(4): 679-686. |
| [7] | Karakousis G, Ye N, Li Z, Chiu SK, Reddy G, Radding CM. The beta protein of phage λ binds preferentially to an intermediate in DNA renaturation. Journal of Molecular Biology, 1998, 276(4): 721-731. DOI:10.1006/jmbi.1997.1573 |
| [8] | Li Z, Karakousis G, Chiu SK, Reddy G, Radding CM. The beta protein of phage λ promotes strand exchange. Journal of Molecular Biology, 1998, 276(4): 733-744. DOI:10.1006/jmbi.1997.1572 |
| [9] | Fu J, Bian XY, Hu S, Wang HL, Huang F, Seibert PM, Plaza A, Xia LQ, Müller R, Stewart AF, Zhang YM. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nature Biotechnology, 2012, 30(5): 440-446. DOI:10.1038/nbt.2183 |
| [10] | Cobb RE, Zhao HM. Direct cloning of large genomic sequences. Nature Biotechnology, 2012, 30(5): 405-406. DOI:10.1038/nbt.2207 |
| [11] | Nawy T. Molecular biology:capturing sequences for bioprospecting. Nature Methods, 2012, 9(6): 532. DOI:10.1038/nmeth.2061 |
| [12] | Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature, 2009, 460(7257): 894-898. DOI:10.1038/nature08187 |
| [13] | Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science, 2011, 333(6040): 348-353. DOI:10.1126/science.1205822 |
| [14] | Wang HH, Kim H, Cong L, Jeong J, Bang D, Church GM. Genome-scale promoter engineering by coselection MAGE. Nature Methods, 2012, 9(6): 591-593. DOI:10.1038/nmeth.1971 |
| [15] | Bird AW, Erler A, Fu J, Hériché JK, Maresca M, Zhang YM, Hyman AA, Stewart AF. High-efficiency counterselection recombineering for site-directed mutagenesis in bacterial artificial chromosomes. Nature Methods, 2011, 9(1): 103-109. DOI:10.1038/nmeth.1803 |
| [16] | Wang HL, Bian XY, Xia LQ, Ding XZ, Müller R, Zhang YM, Fu J, Stewart AF. Improved seamless mutagenesis by recombineering using ccdB for counterselection. Nucleic Acids Research, 2014, 42(5): e37. DOI:10.1093/nar/gkt1339 |
| [17] | Mosberg JA, Gregg CJ, Lajoie MJ, Wang HH, Church GM. Improving lambda red genome engineering in Escherichia coli via rational removal of endogenous nucleases. PLoS One, 2012, 7(9): e44638. DOI:10.1371/journal.pone.0044638 |
| [18] | Thomason LC, Costantino N, Court DL. Examining a DNA replication requirement for bacteriophage λ red-and rac prophage RecET-promoted recombination in Escherichia coli. mBio, 2016, 7(5): e01443-16. |
| [19] | Subramaniam S, Erler A, Fu J, Kranz A, Tang J, Gopalswamy M, Ramakrishnan S, Keller A, Grundmeier G, Müller D, Sattler M, Stewart AF. DNA annealing by Redβ is insufficient for homologous recombination and the additional requirements involve intra-and inter-molecular interactions. Scientific Reports, 2016, 6: 34525. DOI:10.1038/srep34525 |
| [20] |
Hu K, Shi ZX, Wang HL, Feng EL, Huang LY. Study on gene knockout using Red system in Shigella flexneri. Acta Microbiologica Sinica, 2003, 43(6): 740-746.
(in Chinese) 胡堃, 史兆兴, 王恒樑, 冯尔玲, 黄留玉. Red重组系统在痢疾杆菌基因敲除中的应用研究. 微生物学报, 2003, 43(6): 740-746. |
| [21] | Karlinsey JE, Hughes KT. Genetic transplantation:Salmonella enterica serovar Typhimurium as a host to study sigma factor and anti-sigma factor interactions in genetically intractable systems. Journal of Bacteriology, 2006, 188(1): 103-114. DOI:10.1128/JB.188.1.103-114.2006 |
| [22] | Hu SB, Fu J, Huang F, Ding XZ, Stewart AF, Xia LQ, Zhang YM. Genome engineering of Agrobacterium tumefaciens using the lambda Red recombination system. Applied Microbiology and Biotechnology, 2014, 98(5): 2165-2172. DOI:10.1007/s00253-013-5412-x |
| [23] | Kang Y, Norris MH, Wilcox BA, Tuanyok A, Keim PS, Hoang TT. Knockout and pullout recombineering for naturally transformable Burkholderia thailandensis and Burkholderia pseudomallei. Nature Protocols, 2011, 6(8): 1085-1104. DOI:10.1038/nprot.2011.346 |
| [24] | van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nature Methods, 2007, 4(2): 147-152. DOI:10.1038/nmeth996 |
| [25] | van Kessel JC, Hatfull GF. Efficient point mutagenesis in mycobacteria using single-stranded DNA recombineering:characterization of antimycobacterial drug targets. Molecular Microbiology, 2008, 67(5): 1094-1107. DOI:10.1111/mmi.2008.67.issue-5 |
| [26] | van Pijkeren JP, Neoh KM, Sirias D, Findley AS, Britton RA. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered, 2012, 3(4): 209-217. DOI:10.4161/bioe.21049 |
| [27] | Swingle B, Bao ZM, Markel E, Chambers A, Cartinhour S. Recombineering using RecTE from Pseudomonas syringae. Applied and Environmental Microbiology, 2010, 76(15): 4960-4968. DOI:10.1128/AEM.00911-10 |
| [28] | Yin J, Zhu HB, Xia LQ, Ding XZ, Hoffmann T, Hoffmann M, Bian XY, Müller R, Fu J, Stewart AF, Zhang YM. A new recombineering system for Photorhabdus and Xenorhabdus. Nucleic Acids Research, 2015, 43(6): e36. DOI:10.1093/nar/gku1336 |
| [29] | Bode HB Jr, Müller R. The impact of bacterial genomics on natural product research. Angewandte Chemie International Edition, 2005, 44(42): 6828-6846. DOI:10.1002/(ISSN)1521-3773 |
| [30] | Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali J, Linington RG, Fischbach MA. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell, 2014, 158(2): 412-421. DOI:10.1016/j.cell.2014.06.034 |
| [31] | Zarins-Tutt JS, Barberi TT, Gao H, Mearns-Spragg A, Zhang LX, Newman DJ, Goss RJM. Prospecting for new bacterial metabolites:a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Natural Product Reports, 2016, 33(1): 54-72. DOI:10.1039/C5NP00111K |
| [32] | Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nature Reviews Microbiology, 2015, 13(8): 509-523. DOI:10.1038/nrmicro3496 |
| [33] | Nett M. Genome mining:concept and strategies for natural product discovery//Kinghorn AD, Falk H, Kobayashi J. Progress in the Chemistry of Organic Natural Products. Cham: Springer, 2014: 199-245. |
| [34] | Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery:is a renaissance in the making?. Journal of Industrial Microbiology & Biotechnology, 2014, 41(2): 175-184. |
| [35] | Yin J, Wang HL, Li RJ, Ravichandran V, Bian XY, Li AY, Tu Q, Stewart AF, Fu J, Zhang YM. A practical guide to recombineering in Photorhabdus and Xenorhabdus//ffrenchConstant RH. The Molecular Biology of Photorhabdus Bacteria. Cham: Springer, 22017: 195-213. |
| [36] | Ongley SE, Bian XY, Neilan BA, Müller R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Natural Product Reports, 2013, 30(8): 1121-1138. DOI:10.1039/c3np70034h |
| [37] | Wenzel SC, Gross F, Zhang YM, Fu J, Stewart AF, Müller R. Heterologous expression of a myxobacterial natural products assembly line in pseudomonads via Red/ET recombineering. Chemistry & Biology, 2005, 12(3): 349-356. |
| [38] | Perlova O, Fu J, Kuhlmann S, Krug D, Stewart AF, Zhang YM, Müller R. Reconstitution of the myxothiazol biosynthetic gene cluster by Red/ET recombination and heterologous expression in Myxococcus xanthus. Applied and Environmental Microbiology, 2006, 72(12): 7485-7494. DOI:10.1128/AEM.01503-06 |
| [39] | Binz TM, Wenzel SC, Schnell HJ, Bechthold A, Müller R. Heterologous expression and genetic engineering of the phenalinolactone biosynthetic gene cluster by using Red/ET recombineering. ChemBioChem, 2008, 9(3): 447-454. DOI:10.1002/(ISSN)1439-7633 |
| [40] | Hu YF, Phelan VV, Farnet CM, Zazopoulos E, Bachmann BO. Reassembly of anthramycin biosynthetic gene cluster by using recombinogenic cassettes. ChemBioChem, 2008, 9(10): 1603-1608. DOI:10.1002/cbic.v9:10 |
| [41] | Wolpert M, Heide L, Kammerer B, Gust B. Assembly and heterologous expression of the coumermycin A1 gene cluster and production of new derivatives by genetic engineering. ChemBioChem, 2008, 9(4): 603-612. DOI:10.1002/(ISSN)1439-7633 |
| [42] | Fu J, Wenzel SC, Perlova O, Wang JP, Gross F, Tang ZR, Yin YL, Stewart AF, Müller R, Zhang YM. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Research, 2008, 36(17): e113. DOI:10.1093/nar/gkn499 |
| [43] | Bian XY, Tang B, Yu YC, Tu Q, Gross F, Wang HL, Li AY, Fu JM, Shen Y, Li YZ, Stewart AF, Zhao GP, Ding XM, Müller R, Zhang YM. Heterologous production and yield improvement of epothilones in Burkholderiales strain DSM7029. ACS Chemical Biology, 2017, 12(7): 1805-1812. DOI:10.1021/acschembio.7b00097 |
| [44] | Chai Y, Shan SP, Weissman KJ, Hu SB, Zhang YM, Müller R. Heterologous expression and genetic engineering of the tubulysin biosynthetic gene cluster using Red/ET recombineering and inactivation mutagenesis. Chemistry & Biology, 2012, 19(3): 361-371. |
| [45] | Kolinko I, Lohße A, Borg S, Raschdorf O, Jogler C, Tu Q, Pósfai M, Tompa É, Plitzko JM, Brachmann A, Wanner G, Müller, Zhang YM, Schüler D. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nature Nanotechnology, 2014, 9(3): 193-197. DOI:10.1038/nnano.2014.13 |
| [46] | Bian XY, Huang F, Wang HL, Klefisch T, Müller R, Zhang YM. Heterologous production of glidobactins/luminmycins in Escherichia coli Nissle containing the glidobactin biosynthetic gene cluster from Burkholderia DSM7029. ChemBioChem, 2014, 15(15): 2221-2224. DOI:10.1002/cbic.v15.15 |
| [47] | Ongley SE, Bian XY, Zhang YM, Chau R, Gerwick WH, Müller R, Neilan BA. High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chemical Biology, 2013, 8(9): 1888-1893. DOI:10.1021/cb400189j |
| [48] | Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences:synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(44): 15573-15578. DOI:10.1073/pnas.0406911101 |
| [49] | Tu Q, Herrmann J, Hu SB, Raju R, Bian XY, Zhang Y, Müller R M. Genetic engineering and heterologous expression of the disorazol biosynthetic gene cluster via Red/ET recombineering. Scientific Reports, 2016, 6: 21066. DOI:10.1038/srep21066 |
| [50] | Bian XY, Huang F, Stewart AF, Xia LQ, Zhang YM, Müller R. Direct cloning, genetic engineering, and heterologous expression of the syringolin biosynthetic gene cluster in E. coli through Red/ET recombineering. ChemBioChem, 2012, 13(13): 1946-1952. DOI:10.1002/cbic.201200310 |
| [51] | Bian XY, Plaza A, Zhang YM, Müller R. Luminmycins A-C, cryptic natural products from photorhabdus luminescens identified by heterologous expression in Escherichia coli. Journal of Natural Products, 2012, 75(9): 1652-1655. DOI:10.1021/np300444e |
| [52] | Liu QS, Shen QY, Bian XY, Chen HN, Fu J, Wang HL, Lei P, Guo ZH, Chen W, Li DJ, Zhang YM. Simple and rapid direct cloning and heterologous expression of natural product biosynthetic gene cluster in Bacillus subtilis via Red/ET recombineering. Scientific Reports, 2016, 6: 34623. DOI:10.1038/srep34623 |
| [53] | Yin J, Hoffmann M, Bian XY, Tu Q, Yan F, Xia LQ, Ding XZ, Stewart AF, Müller R, Fu J, Zhang YM. Direct cloning and heterologous expression of the salinomycin biosynthetic gene cluster from Streptomyces albus DSM41398 in Streptomyces coelicolor A3(2). Scientific Reports, 2015, 5: 15081. DOI:10.1038/srep15081 |
| [54] | Wang HL, Li Z, Jia RN, Hou Y, Yin J, Bian XY, Li AY, Müller R, Stewart AF, Fu J, Zhang YM. RecET direct cloning and Red αβ recombineering of biosynthetic gene clusters, large operons or single genes for heterologous expression. Nature Protocols, 2016, 11(7): 1175-1190. DOI:10.1038/nprot.2016.054 |
| [55] | Bian XY, Fu J, Plaza A, Herrmann J, Pistorius D, Stewart AF, Zhang YM, Müller R. In vivo evidence for a prodrug activation mechanism during colibactin maturation. ChemBioChem, 2013, 14(10): 1194-1197. DOI:10.1002/cbic.201300208 |
| [56] | Bian XY, Plaza A, Yan F, Zhang YM, Müller R. Rational and efficient site-directed mutagenesis of adenylation domain alters relative yields of luminmide derivatives in vivo. Biotechnology and Bioengineering, 2015, 112(7): 1343-1353. DOI:10.1002/bit.v112.7 |
| [57] | Perlova O, Gerth K, Kuhlmann S, Zhang YM, Müller R. Novel expression hosts for complex secondary metabolite megasynthetases:production of myxochromide in the thermopilic isolate Corallococcus macrosporus GT-2. Microbial Cell Factories, 2009, 8: 1. DOI:10.1186/1475-2859-8-1 |
 2017, Vol. 57
2017, Vol. 57