中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 考桂伟, 邱丽丽, 彭琦, 张杰, 李杰, 宋福平
- Guiwei Kao, Lili Qiu, Qi Peng, Jie Zhang, Jie Li, Fuping Song
- 碱性条件下苏云金芽胞杆菌基础代谢分析
- The analysis of major metabolic pathways in Bacillus thuringiensis under alkaline stress
- 微生物学报, 2016, 56(3): 485-495
- Acta Microbiologica Sinica, 2016, 56(3): 485-495
-
文章历史
- 收稿日期:2015-12-25
- 修回日期:2016-01-21
- 网络出版日期:2016-02-02
2. 东北农业大学生命科学学院,黑龙江 哈尔滨 150030
2. College of Life Sciences, Northeast Agricultural University, Harbin 150030, Heilongjiang Province, China
苏云金芽胞杆菌(Bacillus thuringiensis)是昆虫病原菌,由于其能产生杀虫伴胞晶体(Cry和Cyt毒素蛋白),已成为农业领域广泛应用的微生物农药。在活体实验中B. thuringiensis晶胞混合物毒杀大蜡螟(Galleria mellonella)活性明显高于晶体蛋白[1],说明营养体可能影响晶体蛋白的杀虫效果。鳞翅目昆虫中肠肠道是碱性环境[2],B. thuringiensis在昆虫肠道能否适应生存尚不可知,但碱性环境对B. thuringiensis杀虫效果的发挥至关重要[3]。碱性条件作为胁迫刺激对细菌的生长和繁殖均有较大影响,研究表明在体外实验中,蜡样芽胞杆菌族细胞能够在碱性条件下适应生存[4, 5]。
大肠杆菌Escherichia coli遇到胁迫环境时,一种编码应激蛋白的基因pspA诱导表达[6];在枯草芽胞杆菌Bacillus subtilis中,碱性条件下pspA上调表达,该基因是B. subtilis对碱性条件响应的参考基因[7]。目前关于细菌耐碱适应机制的研究大多围绕细胞表面蛋白及膜离子泵等方面,碱性条件下B. subtilis中δW控制的多个与环境应答有关的基因差异表达[7],其中包括许多表面抗原、双组份信号系统感受蛋白、逆向转运蛋白等;在E. coli中发现,外膜蛋白OmpA及膜结合氧化调节蛋白DsbA在感受外界pH变化时较为敏感[8],说明膜蛋白对细胞适应外界pH变化较为重要。另外,胞内离子浓度变化和离子转运可能也参与了细胞对碱刺激响应[9, 10],高pH条件下乳酸粪肠球菌Streptococcus faecalis负责离子转运的Na+-ATPase的缺失影响了细胞的正常生长[11];在B. subtilis中发现K+(Na+)/H+转运蛋白活动加强[12, 13]。目前除了细菌对碱刺激应答响应的机制研究外,在原核生物中并未发现胞内基础代谢变化影响细胞耐碱能力的相关报道。
本实验室前期研究发现在液体培养B. thuringiensis菌株时加入NaOH,之后培养基pH逐渐下降;B. thuringiensis比B. subtilis具有更强的碱适应能力,B. thuringiensis胞外pH下降至8.9-9.1时菌体即可恢复生长,而B. subtilis菌株需胞外pH下降至8.2-8.4才能恢复生长[14],说明B. thuringiensis适应碱性环境可能具有自身的特点。本文通过探索B. thuringiensis对碱刺激条件的响应,利用表达谱芯片研究B. thuringiensis HD73菌株的全基因组表达情况,重点分析了碱刺激胁迫下B. thuringiensis HD73基础代谢变化可能对细胞耐碱能力产生的影响。
1 材料和方法 1.1 菌种和培养条件供试菌株为本实验室保存的B. thuringiensis serovar. kurstaki HD73菌株,在LB培养基中按照1%接种量接种,30 ℃、220 r/min条件下培养。
1.2 半定量RT-PCR、实时荧光定量PCRHD73菌株在LB培养基中培养至对数生长中期,在处理组菌体中加入终浓度为28 mmol/L的NaOH,对照组菌体不进行加碱处理,继续培养10 min后快速取样,低温离心弃上清,沉淀立即用预冷的TRIzol重悬,RNA提取按照Qiagen Easy RNA kit说明。RNA纯化后电泳检测,-70 ℃保存备用。以纯化后的RNA为模板合成cDNA,RT-PCR试剂盒为Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo公司),实时荧光定量qRT-PCR试剂盒为Go Taq qPCR Master Mix (Promega公司),所用引物见表 1。
| Primer names | Sequences (5′→3′) |
| 16S rRNA5′ | ATCTTCCGCAATGGACGAAAGTC |
| 16S rRNA3′ | GGTCTTGCAGCTCTTTGTACCGT |
| pspA5′ | CCGCCAGCAGTACGAAATAA |
| pspA3′ | GCCGCATTTAACCTTGAAACA |
实验芯片杂交前采用Agilent表达谱芯片配套试剂盒和标准操作流程对样品total RNA中的mRNA进行放大和标记,并用RNeasy mini kit纯化标记产物cRNA。随后取600 ng cRNA进行芯片杂交,在滚动杂交炉(Hybridization Oven)中65 ℃、10 r/min滚动杂交17 h,在洗缸中进行洗片处理。最后对完成杂交的芯片进行Agilent Microarray Scanner扫描,用Feature Extraction software 10.7读取数据并通过Gene Spring Software 11.0完成归一化处理。
1.4 GO富集分析和KEGG分析HD73基因组[15]和对应蛋白序列均下载于NCBI数据库。为了获取更精确的GO注释信息,利用Blast2GO软件对HD73的蛋白序列进行比对和注释。HD73基因以及基础代谢途径注释来自于KEGG数据库(http://www.genome.jp/kegg/)。后续富集分析采用本地编写的Perl程序完成。
2 结果和分析 2.1 pspA基因在碱性条件下诱导表达在HD73基因组中发现了B. subtilis碱刺激响应参考基因pspA的同源基因HD73_1073,蛋白序列比对Identity约为42.9%,以该基因表达情况作为衡量B. thuringiensis细胞对碱刺激响应的标准。对处理组和对照组所提取的RNA进行半定量RT-PCR实验,发现细胞对数生长中期时加入终浓度为28 mmol/L的NaOH并诱导培养10 min后,在内参基因16S rRNA表达量相同的情况下,pspA基因的转录表达发生明显变化(图 1-A)。利用qRT-PCR验证同样条件下pspA差异表达情况,碱性处理组的pspA表达量明显上调(图 1-B)。因此将上述时期作为碱刺激处理条件,提取RNA进行基因芯片杂交。对NaOH处理的实验组和不采用NaOH处理的对照组分别设置3个生物学重复。

|
| 图 1. B. thuringiensis HD73响应碱性条件的确定及验证 Figure 1. Expression of pspA was confirmed by RT-PCR and qRT-PCR. A: Semi-quantitative RT-PCR analysis of the transcription of pspA. The sample treated with NaOH was labeled “+”, and sample untreated was labeled “–”. B: qRT-PCR analysis of the transcription of pspA. The sample treated with NaOH was labeled “T”, and sample untreated was labeled “C”. The bars represent the mean±S.D (n=3). |
根据Scanner扫描发现共检测到5436个基因表达信号,约占HD73基因总数的87%。为了保证数据可靠性,采用严格的域值(Fold change>2.00或Fold change<0.50,P<0.05)筛选差异表达基因。与对照组相比,碱处理后约有1400个基因存在表达差异,其中47% (662个)基因上调表达,53% (739个)基因下调表达,有432个基因为未知功能或功能不明确的基因。
Gene Ontology (简称GO)和KEGG 途径的富集分析是系统性描述表达谱数据变化概况和趋势的工具,能够为进一步挖掘利用表达谱信息提供分析基础。以P值小于0.05 (Chi-squared test)为阈值对表达谱进行GO富集分析,发现细胞的物质代谢、有机物合成、氧化还原反应等多个途径明显上调富集(图 2)。在KEGG富集分析中,参与细胞基础代谢途径的基因明显上调,包括碳代谢循环、脂肪酸代谢及氨基酸的合成与转运。由于下调基因存在较多未知功能蛋白,分析中并未出现明显抑制富集的途径。

|
| 图 2 碱性条件下上调基因GO富集情况 Figure 2. GO enrichments of up-regulated genes under alkaline stress. |
综合以上分析结果可知,B. thuringiensis在碱性条件下的基因表达情况并不是单一、独立的改变,而是在细胞基础代谢及信号调控等方面均存在系统性差异。由于基础代谢对于细胞生长是不可或缺的过程,以下重点对碱性条件下基础代谢途径变化进行分析。
2.3 碱性条件对糖酵解途径的影响糖酵解是为细胞提供能量的主要途径,有研究报道细菌遇到胁迫条件时多糖代谢明显加强[16],而本研究也发现这一现象。碱刺激胁迫下细胞内糖酵解途径中至少19个编码重要酶类的基因明显上调,涉及到糖酵解多数反应过程(图 3-A),而糖异生的过程并没有明显差异变化,仅有fruK (Gene ID: HD73_4002)出现抑制表达,说明此时细胞加快对葡萄糖的利用,糖酵解途径显著加强,丙酮酸激酶编码基因上调表达2倍,促使细胞产生更多丙酮酸。然而在丙酮酸的代谢途径中,催化丙酮酸转换为乙酰-CoA的丙酮酸脱氢酶并未出现差异表达。从图 3中可以看到参与途径8中的乳酸脱氢酶基因ldh2 (Gene ID: HD73_5189)显著上调,诱导倍数高达35倍。随着糖酵解代谢增强而不断积累的丙酮酸可能并没有及时转换为乙酰-CoA进入TCA循环,而是以合成乳酸的方式完成一部分能量代谢。丙酮酸转化为乳酸能够为胞内提供更多的有机酸类物质以调节胞内pH,且产生的NAD+能够持续为糖酵解反应提供底物,以此保证糖酵解途径中ATP的生成。
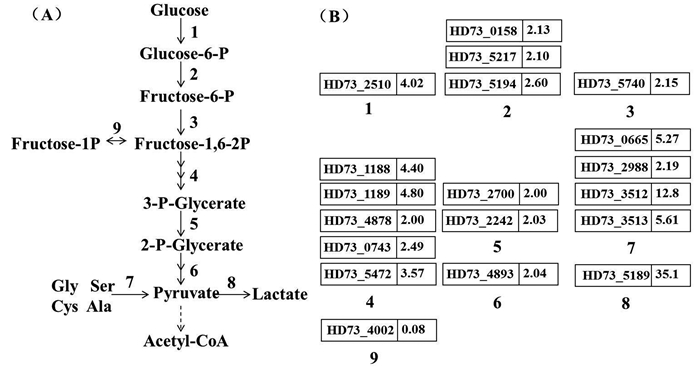
|
| 图 3. 碱性条件下糖酵解途径及基因差异表达情况 Figure 3. Glycolytic pathway and involved differential expressed genes of B. thuringiensis under alkaline stress. A: The main steps in glycolytic pathway were shown. Numbers 1–9 represent different enzymes. 1: Glucokinase; 2: Glucose phosphate isomerase; 3: Phosphofructokinase; 4: Aldolase, Triose phosphofructokinase, Glyceraldehyde phosphate dehydrogenase, Phosphoglycerate kinase; 5: Phosphoglyceromutase; 6: Pyruvate kinase; 7: Serine dehydratase, Alanine dehydrogenase, Glyoxalase; 8: Lactate dehydrogenase; 9: Phosphofructokinase. B: The fold changes of involved genes of B. thuringiensis under alkaline stress. |
从图 4中可知参与乙酰-CoA合成α-酮戊二酸的很多酶蛋白编码基因未出现差异表达,与上述推测一致。但在α-酮戊二酸合成苹果酸的过程中,包括sucB (Gene ID: HD73_1488)、sucC (Gene ID: HD73_4119)、fumA (Gene ID: HD73_0553)在内的12个酶促基因明显上调,而由苹果酸转化为草酰乙酸的苹果酸脱氢酶编码基因未出现差异表达,说明在碱性条件下苹果酸的合成与积累明显加强,且产生的苹果酸并没有通过转化为草酰乙酸而降解。从代谢途径13 (图 4)中发现,合成α-酮戊二酸的氨基转移酶表现诱导上调,即谷氨酸的降解代谢为细胞提供了进行TCA循环的底物α-酮戊二酸,以上途径帮助细胞合成更多苹果酸。积累的苹果酸可能和乳酸发挥类似作用,这对于细胞响应碱刺激环境胁迫,调节细胞生存微环境中的酸碱平衡具有重要意义。
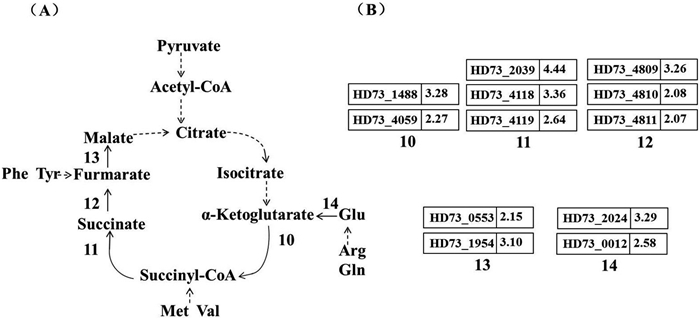
|
| 图 4. 碱性条件下HD73的TCA循环途径及基因差异表达情况 Figure 4. TCA cycle and involved differential expressed genes of B. thuringiensis under alkaline stress. A: The main steps in TCA cycle were shown. Numbers 10–14 represent different enzymes. 10: α-Ketoglutarate dehydrogenase, Oxoglutarate dehydrogenase; 11: Succinyl-CoA synthetase, Succinate-CoA transferase; 12: Succinate dehydrogenase; 13: Fumarase; 14: Glutamine dehydrogenase. B: The fold changes of involved genes of B. thuringiensis under alkaline stress. |
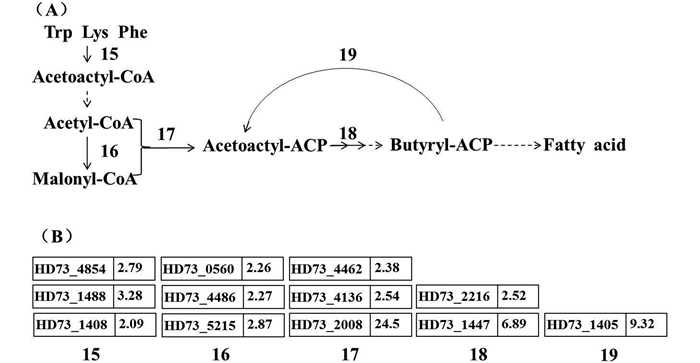
色氨酸、赖氨酸、苯丙氨酸能够为脂肪酸合成提供乙酰乙酰-CoA,乙酰乙酰-CoA通过脱酰基作用可变为乙酰-CoA。乙酰-CoA和丙二酸单酰-CoA是脂肪酸合成的底物。乙酰-CoA羧化酶能将乙酰-CoA转化为丙二酸单酰-CoA,在原核生物中乙酰-CoA羧化酶由生物素羧基载体蛋白在内的多个蛋白复合体组成[17]。芯片分析发现与合成生物素相关的bio家族基因均诱导上调。在脂肪酸合成的起始阶段中,多个合成酶基因诱导上调,脂肪酸合成速度加快(图 5)。与脂肪酸氧化代谢相关的脂酰-CoA脱氢酶的编码基因HD73_3476、脂肪酸去饱和酶编码基因HD73_2987、脂肪酸激酶编码基因HD73_4466均上调表达2倍以上,在碱性条件下脂肪酸合成及代谢反应均明显加强。
|
| 图 5. 碱性条件下HD73中脂肪酸合成及基因差异表达情况 Figure 5. Initial steps of fatty acid synthesis pathway and involved differential expressed genes of B. thuringiensis under alkaline stress. A: The initial steps of fatty acid synthesis pathway were shown. Numbers 15–19 represent different enzymes. 15: Trp, PheS, LysS; 16: Acetyl-CoA carboxylase; 17: Lipoamide acyltransferase, Acyl-carrier-protein synthase; 18: Enoyl-reductase, Acyl-CoA thioester hydrolase; 19: Oxoacyl-ACP synthase. B: The fold changes of involved genes of B. thuringiensis under alkaline stress. |
上文提到在碱性条件下细胞存在丙酮酸大量合成的情况,根据氨基酸代谢变化发现:可形成丙酮酸的谷丙转氨酶、半胱氨酸合成酶、丝氨酸-tRNA合成酶的多个编码基因上调,说明这些氨基酸的合成明显增强,并可能与丙酮酸的代谢相关,从而实现对外界刺激的适应。
α-酮戊二酸是连接氨基酸代谢和TCA循环的重要中枢分子,能够形成α-酮戊二酸的氨基酸有精氨酸、组氨酸、谷氨酸[18],在芯片数据中负责此类氨基酸降解的诸多基因均明显上调。甲硫氨酸、缬氨酸与琥珀酰-CoA的合成密切相关,其合成酶基因均诱导上调。延胡索酸是合成苹果酸的直接底物,对于苹果酸的合成至关重要,而与延胡索酸有关的苯丙氨酸和酪氨酸的代谢也明显增强。
亮氨酸tRNA合成酶基因、缬氨酸tRNA合成酶基因均诱导表达(表 2),这些氨基酸能够为支链脂肪酸的合成提供前体物质。综上,与糖代谢和脂肪酸代谢途径相关的氨基酸的合成代谢途径都有明显增强,说明细胞可能通过三大类物质的互相转化,保证胞内有机酸合成,为细胞抵抗碱性刺激提供物质基础。
| Related pathway | Gene ID | Fold change | P-value | Gene product |
| Ketoglutaric acid biosynthesis | HD73_4433 | 3.37 | 0.049 | Arginine bifunctional protein |
| HD73_0012 | 2.58 | 0.014 | Glutamine amidotransferase | |
| HD73_0984 | 2.50 | 0.044 | Glutamate racemase | |
| HD73_5377 | 5.01 | 0.007 | L-proline dehydrogenase | |
| Succinyl-CoA biosynthesis | HD73_0036 | 3.10 | 0.010 | Methionyl-tRNA synthetase |
| HD73_5073 | 2.41 | 0.004 | Adenosylmethionine synthetase | |
| HD73_2042 | 3.62 | 0.008 | Methylthioadenosine deaminase | |
| HD73_1436 | 3.49 | 0.030 | Methyltransferase | |
| HD73_4765 | 2.64 | 0.001 | Valyl-tRNA synthetase | |
| Furmarate biosynthesis | HD73_4854 | 2.79 | 0.001 | Phenylalanyl-tRNA synthetase |
| HD73_1605 | 2.49 | 0.002 | Aminoacylase | |
| Oxaloacetate biosynthesis | HD73_4172 | 10.5 | 0.006 | Dihydroorotase |
| HD73_0685 | 4.33 | 0.029 | Aspartate ammonia-lyase | |
| HD73_4173 | 8.56 | 0.008 | Aspartate carbamoyltransferase | |
| Fatty acid biosynthesis | HD73_4486 | 2.10 | 0.001 | Seryl-tRNA synthetase |
| HD73_5048 | 2.33 | 0.001 | Leucyl-tRNA synthetase | |
| HD73_3511 | 7.75 | 0.046 | Serine transporter | |
| HD73_3513 | 12.8 | 0.030 | L-serine dehydratase | |
| HD73_4486 | 2.27 | 0.002 | Acetyl-CoA carboxylase biotin carboxylase |
HD73在应答碱性胁迫条件时在细胞膜组分、转录调控、物质转运系统等方面均存在诸多差异表达基因,至少15个细胞膜组分基因诱导表达(表 3),其中包括已在E. coli耐碱适应中研究过的膜蛋白基因ompA[8]的同源基因romA (Gene ID: HD73_3311),有至少18个膜组分基因抑制表达(表 4)。在转录调控因子的表达情况中发现一个δ因子编码基因(Gene ID: HD73_5474)上调表达4.94倍,另外有Crp、LacI、LysR、TetR等12个转录调控因子的编码基因上调表达,6个与双组份信号系统调节因子编码基因抑制表达。
| Function | Gene ID | Fold change | P-value | Gene product |
| Membrane component | HD73_3311 | 2.68 | 0.035 | Outer membrane protein romA |
| HD73_5566 | 13.17 | 0.021 | UPF0324 membrane protein | |
| HD73_3039 | 4.59 | 0.036 | Integral membrane protein TerC | |
| HD73_2328 | 3.79 | 0.009 | Putative Membrane Associated Protein | |
| HD73_2180 | 2.68 | 0.011 | Putative membrane protein | |
| HD73_0451 | 3.38 | 0.044 | Mandelate racemase/Muconate | |
| HD73_2850 | 2.07 | 0.018 | Cell wall anchor domain-containing protein | |
| HD73_2987 | 8.28 | 0.012 | Fatty acid desaturase (Membrane bound) | |
| HD73_4342 | 2.24 | 0.017 | Beta-lactamase | |
| HD73_0342 | 2.07 | 0.018 | Phosphoribosylamine--glycine ligase | |
| HD73_4083 | 3.00 | 0.006 | Dihydrodipicolinate synthase | |
| HD73_4680 | 4.09 | 0.030 | Cystathionine gamma-synthase | |
| HD73_2476 | 5.11 | 0.029 | Hypothetical protein | |
| HD73_2479 | 2.54 | 0.001 | Hypothetical protein | |
| HD73_2897 | 4.09 | 0.005 | Hypothetical protein | |
| Transcription factor | HD73_0493 | 21.70 | 0.027 | Crp family transcriptional regulator |
| HD73_1240 | 11.31 | 0.003 | Transcriptional regulator, LacI | |
| HD73_5474 | 4.94 | 0.009 | RNA polymerase sigma-54 factor | |
| HD73_5530 | 2.11 | 0.031 | Rha family phage regulatory protein | |
| HD73_4979 | 2.64 | 0.039 | Response regulator receiver domain protein | |
| HD73_2097 | 2.70 | 0.012 | Transcriptional regulator, LysR | |
| HD73_2836 | 2.70 | 0.044 | Transcriptional regulator, TetR | |
| HD73_2314 | 2.59 | 0.004 | Transcriptional regulator, GntR | |
| HD73_5854 | 80.70 | 0.001 | Murein hydrolase exporter | |
| HD73_2788 | 2.25 | 0.002 | Hypothetical protein | |
| HD73_1239 | 10.40 | 0.002 | Hypothetical protein | |
| HD73_5853 | 94.80 | 0.001 | Hypothetical protein |
| Function | Gene ID | Fold change | P-value | Gene product |
| Membrane component | HD73_1419 | 0.15 | 0.001 | Putative membrane protein |
| HD73_2214 | 0.19 | 0.016 | Putative membrane protein | |
| HD73_4460 | 0.21 | 0.020 | Putative membrane protein | |
| HD73_5464 | 0.26 | 0.006 | Membrane protein, putative | |
| HD73_2313 | 0.27 | 0.031 | Putative Membrane Spanning Protein | |
| HD73_4874 | 0.30 | 0.047 | Membrane attachment protein | |
| HD73_2141 | 0.35 | 0.001 | MgtC/SapB family membrane protein | |
| HD73_0351 | 0.48 | 0.022 | Putative membrane spanning protein | |
| HD73_2108 | 0.15 | 0.149 | DegT/DnrJ/EryC1/StrS | |
| HD73_2209 | 0.40 | 0.041 | Hypothetical protein | |
| HD73_4686 | 0.38 | 0.002 | Hypothetical protein | |
| HD73_1886 | 0.35 | 0.001 | Hypothetical protein | |
| HD73_3447 | 0.21 | 0.015 | Sortase | |
| HD73_5605 | 0.33 | 0.002 | Teichoic acid linkage unit synthesis | |
| HD73_5841 | 0.22 | 0.008 | Glycosyltransferase involved in cell wall biogenesis | |
| HD73_3575 | 0.28 | 0.021 | LPXTG-domain-containing protein cell wall anchor domain | |
| HD73_0603 | 0.24 | 0.040 | Daunorubicin resistance transmembrane protein | |
| HD73_5779 | 0.48 | 0.012 | Membrane protein PfoR | |
| Two-component system | HD73_5882 | 0.50 | 0.023 | Two-component sensor kinase yycG |
| HD73_0642 | 0.12 | 0.045 | Two-component response regulator | |
| HD73_5397 | 0.31 | 0.001 | Two-component response regulator yanRB | |
| HD73_5163 | 0.33 | 0.003 | Two component regulator, winged helix | |
| HD73_1669 | 0.41 | 0.009 | Two-component response regulator yvqC | |
| HD73_2137 | 0.41 | 0.012 | Two component transcriptional regulator | |
| HD73_1668 | 0.28 | 0.002 | Sensor histidine kinase | |
| HD73_0460 | 0.38 | 0.036 | Methylthioribose kinase | |
| HD73_4343 | 0.16 | 0.045 | Sensory box/GGDEF family protein | |
| HD73_5707 | 0.17 | 0.028 | Sensory box/GGDEF |
细菌耐碱适应是一个较为复杂的过程。细胞在感受到酸碱胁迫刺激,完成胞内外信号传导应答后,将快速调整自身基因表达来适应外界胁迫环境的变化,其中通过调节pH稳态是一种较为重要的适应方式[19]。有研究表明,碱性条件下胞质内pH下降,细胞不需要保持胞内中性环境即能维持生长[20]。在真核生物中发现,碱刺激下细胞合成苹果酸的能力增强[21],但在原核生物中并未发现关于碱刺激下胞内酸性大分子物质合成情况的报道。根据表达谱芯片结果,本研究发现在碱刺激下HD73胞内苹果酸在合成途径上的变化,苹果酸不仅由通常的碳代谢模式合成,而且可能通过加强α-酮戊二酸的合成和代谢逐渐积累。除此之外,细胞内糖酵解途径产生的丙酮酸及丙酮酸代谢合成乳酸的能力均明显增强,这说明B. thuringiensis不仅存在苹果酸大量合成的情况,同时也可能通过合成丙酮酸、乳酸等多种酸类物质来适应外界高pH。
在其他基础代谢中,细胞脂肪酸和氨基酸的合成与代谢均有不同程度的加强,尤其与苹果酸合成途径相关的氨基酸合成与代谢的基因多数诱导显著。碱刺激条件使E. coli的精氨酸和谷氨酸降解代谢发生变化[22],本研究结果中也发现这一情况,除此之外还有很多与苹果酸合成相关的氨基酸基因诱导表达,这说明碱刺激胁迫促使细胞多种不同途径同时差异变化,以协调酸类物质的合成。综上,尽管在B. thuringiensis中可能还存在着其他的耐碱机制,但是本文中发现了B. thuringiensis在酸类物质的合成模式、诸多基础代谢途径的变化,为进一步分析和探究菌体耐碱的作用机理提供了新的思路。
| [1] | Fedhila S, Buisson C, Dussurget O, Serror P, Glomski IJ, Liehl P, Lereclus D, Nielsen-LeRoux C. Comparative analysis of the virulence of invertebrate and mammalian pathogenic bacteria in the oral insect infection model Galleria mellonella. Journal of Invertebrate Pathology, 2010, 103(1): 24–29. |
| [2] | Berenbaum M. Adaptive significance of midgut pH in larval Lepidoptera. American Naturalist, 1980: 138–146. |
| [3] | Knowles BH. Mechanism of action of Bacillus thuringiensis insecticidal d-endotoxins. Advances in Insect Physiology, 1994, 24: 275–308. |
| [4] | Cao M, Kobel PA, Morshedi MM, Wu MFW, Paddon C, Helmann JD. Defining the Bacillus subtilis δW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. Journal of Molecular Biology, 2002, 316(3): 443–457. |
| [5] | Flahaut S, Hartke A, Giard J-C, Auffray Y. Alkaline stress response in Enterococcus faecalis: adaptation, cross-protection, and changes in protein synthesis. Applied and Environmental Microbiology, 1997, 63(2): 812–814. |
| [6] | Kleerebezem M, Crielaard W, Tommassen J. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. The EMBO Journal, 1996, 15(1): 162. |
| [7] | Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis δW regulon. Molecular Microbiology, 2001, 41(1): 59–71. |
| [8] | Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. Journal of Bacteriology, 2002, 184(15): 4246–4258. |
| [9] | Kawano M, Igarashi K, Kakinuma Y. Isolation of Enterococcus hirae mutant deficient in low-affinity potassium uptake at alkaline pH. Bioscience, Biotechnology, and Biochemistry, 2002, 66(7): 1597–1600. |
| [10] | Trchounian A, Kobayashi H. Relationship of K+-uptaking system with H+-translocating ATPase in Enterococcus hirae, grown at a high or low alkaline pH. Current Microbiology, 1998, 36(2): 114–118. |
| [11] | Kakinuma Y, Igarashi K. Mutants of Streptococcus faecalis sensitive to alkaline pH lack Na (+)-ATPase. Journal of Bacteriology, 1990, 172(4): 1732–1735. |
| [12] | Fujisawa M, Wada Y, Ito M. Modulation of the K+ efflux activity of Bacillus subtilis YhaU by YhaT and the C-terminal region of YhaS. FEMS Microbiology Letters, 2004, 231(2): 211–217. |
| [13] | Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C–125. Molecular Microbiology, 1994, 14(5): 939–946. |
| [14] | Qiu LL, Peng Q, Qu N, Liu CX, Li J, Zhang J, Song FP. Effect of clpP disruption on alkaline sensitivity of Bacillus thuringiensis. Acta Microbiologica Sinica, 2013, 53(6): 615–622. (in Chinese)邱丽丽, 彭琦, 曲宁, 刘春霞, 李杰, 张杰, 宋福平. 苏云金芽胞杆菌clpP基因缺失对碱刺激敏感性的影响. 微生物学报, 2013, 53(6): 615–622. |
| [15] | Liu G, Song L, Shu C, Wang P, Deng C, Peng Q, Lereclus D, Wang X, Huang D, Zhang J. Complete genome sequence of Bacillus thuringiensis subsp. kurstaki strain HD73. Genome Announcements, 2013, 1(2): e00080–00013. |
| [16] | Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Current Issues in Molecular Biology, 2005, 7(1): 95–108. |
| [17] | Rodrigues TE, Gerhardt E, Oliveira MA, Chubatsu LS, Pedrosa FO, Souza EM, Souza GA, Müller‐Santos M, Huergo LF. Search for novel targets of the PII signal transduction protein in Bacteria identifies the BCCP component of acetyl‐CoA carboxylase as a PII binding partner. Molecular Microbiology, 2014, 91(4): 751–761. |
| [18] | Fernández M, Zúñiga M. Amino acid catabolic pathways of lactic acid bacteria. Critical Reviews in Microbiology, 2006, 32(3): 155–183. |
| [19] | Slonczewski JL, Rosen BP, Alger JR, Macnab RM. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proceedings of the National Academy of Sciences, 1981, 78(10): 6271–6275. |
| [20] | Mugikura S, Nishikawa M, Igarashi K, Kobayashi H. Maintenance of a neutral cytoplasmic pH is not obligatory for growth of Escherichia coli and Streptococcus faecalis at an alkaline pH. Journal of Biochemistry, 1990, 108(1): 86–91. |
| [21] | Žárský V. Signal transduction: GABA receptor found in plants. Nature Plants, 2015, 1(8). |
| [22] | Yohannes E, Barnhart DM, Slonczewski JL. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli K-12. Journal of Bacteriology, 2004, 186(1): 192–199. |
 2016, Vol. 56
2016, Vol. 56




