中国科学院微生物研究所,中国微生物学会,中国菌物学会
文章信息
- 熊天庆, 王登莉, 谭百宏, 姜文华, 代高月, 赵慧, 李树蕾, 李艳超. 2015
- Tianqing Xiong, Dengli Wang, Baihong Tan, Wenhua Jiang, Gaoyue Dai, Hui Zhao, Shulei Li, Yanchao Li. 2015
- 人类及小鼠某些细胞系培养中苯基杆菌的污染
- Contamination of Phenylobacterium in several human and murine cell cultures
- 微生物学报, 2015, 55(2): 176-186
- Acta Microbiologica Sinica, 2015, 55(2): 176-186
-
文章历史
- 收稿日期:2014-04-24
- 修回日期:2014-06-15
2. 冈山大学大学院医齿药学综合研究科药理学系,日本 冈山县 700-8530;
3. 吉林大学白求恩医学部基础医学院基础医学实验中心,吉林 长春 130021
2. Department of Pharmacology, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama 700-8530, Japan;
3. Laboratory Teaching Center of Basic Medicine, Norman Bethune Health Science Center of Jilin University, Changchun 130021, Jilin Province, China
Culture contamination is a common problem for most researchers,and often results in failure of experiments,and loss of time and money. The contamination may not be easy to identify,mainly due to the fact that the identification is time-consuming and often deviates from the initial goals of researchers. However,if the nature of contamination remains unknown,erroneous explanations for experimental results may be incurred due to the potential responses of the host cells to the infection.
In our laboratory,a rod-shaped microorganism has contaminated many cell lines. It resembles cell debris,but is motile,and accumulates slowly in the media and in the cell bodies of cultured cells. In most cases,the cells appear healthy,and the media are neither prematurely acidic nor grossly turbid. The contamination often subsides somehow,but breaks out overnight. Discussion of similar culture contamination can be found in many online science blogs. The researchers and students on these blogs described a swimming rod-shaped microorganism in their cultures,which has the same features to that in our laboratory.
Here we reported the characterization of such cell culture contamination by 16S rRNA sequence analysis,as well as the information regarding antibiotic resistance and treatment effectiveness. One potential problem of such bacterial contamination is unsuccessful resuscitation of thawed cells. In this study we reported a simple resuscitating method which remarkably promoted the survival of the thawed cells.
1 Materials and Methods 1.1 Cell CultureHuman neuroblastoma SH-SY5Y cell line was purchased from ATCC,and was passaged in the laboratory for less than 6 months. Mouse macrophage cell line (RAW 264.7) was a gift from Dr. Q Liu (Dept. Immunology,Jilin Univ.,China). This cell line was established from the ascites of a male mouse with a tumor induced by intraperitoneal injection of Abselon Leukemia Virus[1]. It has receptors for immunoglobulin and produces lysozymes,but is negative for surface immunoglobulin,Ia and Thy-1.2. Murine Lewis lung carcinoma cell line was obtained from ATCC,which was established from Lewis lung carcinoma,and expressed H-2b antigen[2]. Mouse neuroblastoma N2a cells were established from a spontaneous tumor of an albino mouse[3]. The cells expressed H-2 antigen (a haplotype),and produced large quantities of microtubular protein[4].
These cells were routinely grown in Dulbecco’s modified Eagle’s Medium (DMEM,Gibco Invitrogen,Carsbad,C.A.,U.S.A.) supplemented with 10% Gamma-irradiated fetal bovine serum (Biological industries,Kibbutz Beit haemek 25115,Israel). The cells were incubated at 37℃ in a humidified atmosphere of 95% air/5% CO2 with the media changed every 1-2 days.
The cells were photographed every day with an inverted microscope (Olympus IX 71,Tokyo,Japan). The bacterium was video-recorded for 10-20 s with a 40×objective lens using the software Motic images Advanced 3.2 (Motic China Group Co. LTD.). Swimming trajectories were analyzed using ImageJ Particle Tracker plugin[5] and custom MATLAB scripts (The Mathworks,Natick,M.A.). The speed was then calculated from these trajectories by calculating the average distance traveled by the bacterium between consecutive frames,and each measurement was repeated at least three times.
1.2 Morphological observations with fluorescence and electron microscopesThe cell lines seeded on coverslips were incubated with 300 nmol/L 4',6-Diamidino-2-phenylindole (DAPI,Gibco Invitrogen) in 0.1 mol/L phosphate buffer (PB) for 5 minutes,and then with 10 nmol/L 3,3'-Dihexyloxacarbocyanine Iodide (DiOC6(3),FluoProbe,interchim) in PB for 15 minutes. Culture supernatants from different contamination stages were smeared on the glass slides,and processed for Gram staining and acid-fast staining.
The culture supernatants were applied to copper meshes,stained with 2% phosphotungstic acid for 1-2 min,and examined under a high resolution transmission electron microscope (JEOL JEM 1200EXⅡ). The short and long diameters of the bacterium were measured on the electron micrographs using the ImageJ program.
1.3 Bacterial culture,DNA isolation and 16S rRNA sequencingThe bacterium was cultured either in liquid media such as DMEM or on Luria-Bertani (LB) agar. After 3 to 5 days,the bacteria from LB agar and DMEM were collected respectively for genomic DNA isolation using a SK 1201-UNIQ-10 genomic DNA isolation Kit (Sangon Biological Engineering Tech.,ShangHai,China). About 0.3 μg of purified DNA was subjected to polymerase chain reaction (PCR) amplification of 16S rRNA with the primers 7f (5'-CAGAGTTTGATCCTGGCT-3') and 1540r (5'-AGGAGGTGATCCAGCCGCA-3'). The PCR was run using the following cycling parameters: 5 min 94℃; 35 cycles of 30 s 94℃,35 s 55℃,1min 72℃,with a final elongation step of 8 min 72℃.
PCR products were visualized on 1% agarose gel,isolated with UNIQ-10 DNA retrieval kit (Sangon Biological Engineering Tech.,Shanghai,China),and processed for sequencing with ABI 3730 DNA analyzer (Applied Biosystems,Foster City,CA). The 16S rRNA sequence was compared with the known sequences in the GenBank/EMBL/DDBJ databases.
1.4 Antibiotic treatmentsThe bacterium was plated onto LB agar containing 5% sheep erythrocytes at 37℃,and the bacterial inhibition ring tests for standard drug sensitivity were carried out. Paper discs containing different types and amounts of antibiotics were added to the bacterial culture. After 24 hours,the culture was examined to see which antibiotic was the most effective in destroying or inhibiting the bacterium according to the width of the inhibition ring.
Based on the drug sensitivity test and on the data reported previously for Phenylobacterium[6, 7, 8] and intracellular bacteria[9, 10],a variety of antibiotics of various concentrations (Table1) were added into the contaminated cell lines with the media changed every 1-2 days. The freshly prepared complete medium was added as a negative control. The contaminated supernatant without any treatment was used as a positive control. All analyses were performed at least in triplicate,and each experiment was repeated for at least 3 times.
| Pharmaceuticals | Dose(μg/mL) |
| penicillin | 60-6000 |
| streptomycin | 100-10000 |
| ciprofloxacin | 10-100 |
| levofloxacin | 10-100 |
| piperacillin | 10-100 |
| sulfadiazine | 10-400 |
| acriflavine hydrochloride | 0.44-22 |
| oxytetracycline | 10-400 |
| amphotericin B | 0.5-5.0 |
| azithromycin | 1.0-200 |
| gentamicin | 5-200 |
| diminazene aceturate | 10-400 |
| p-aminosalicilic acid | 25-200 |
| ethionamide | 25-200 |
| enrofloxacin | 50-400 |
| imipenem | 100-400 |
During experiments,we found that the bacterium frequently became indiscernible in some culture vessels,and the cells looked healthy again. These cells were selected and routinely frozen in 90% FBS supplemented with 10% DMSO at-80℃. Meanwhile the culture supernatants were also collected and kept at-80℃ for future resuscitation.
One year later,the cryostored cells were thawed either in the culture supernatants collected before or in freshly prepared media. The number of resuscitated cells was counted by using the ImageJ program,and presented as Mean±standard deviation. Statistical analyses were done using Student’s t test or one-way ANOVA with significance at values of P<0.05.
1.6 The bacterial infection in SH-SY5Y cellsIn order to further observe the relationship between the bacterium and host cells,the bacterium was adjusted to 108 CFU/mL with DMEM,and used to infect healthy SH-SY5Y cells. When the cells grew to approximately 70%-90% confluence,about 1μL,10μL,20μL of the bacterial solutions were added to the culture medium,respectively,with six replicate wells for each concentration. After that,the cultured cells were examined at 0 h,24 h and 48 h,and the number of survived cells was counted by using the ImageJ program,and presented as Mean ±standard deviation. Statistical analyses were done by using one-way ANOVA with significance set at P<0.05.
2 Results 2.1 Light and fluorescence microscopic observationsThe morphology and motility of the bacterium in the cultures was examined by using an inverted microscope. Based upon the morphological observations,the growth of the unknown bacterium was artificially divided into 3 stages for description convenience.
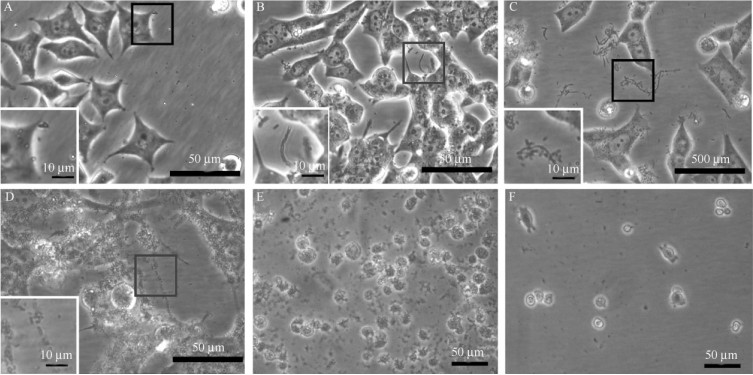
In the early stage,the cell lines generally grew well,while the bacteria were few,and were often easily to be neglected (Figure1-A). They were rod-shaped,generally no more than 2.0 μm in length,and most of them adhered to the surface of the host cells (inset in Figure1-C). A few of them were vibrating in situ,or swimming around for a very short distance with a speed of about 0.46 μm/s. The blue-fluorescent DAPI preferentially stains nuclear DNAs with little or no cytoplasmic labeling. However,both the nuclei and the cytoplasm of the contaminated cells were strongly stained by DAPI,suggesting that the cytoplasm might contain foreign DNAs from unknown bacteria.
|
| Figure. 1 The swimming rod-like structures occurring in the cell culture. Scale bars: 50 μm; insets, 10 μm. A-D: the different growth stages of rod-like structures in SH-SY5Y cell culture. The insets in A-D are enlarged in each panel, respectively. A: In the early stage, the cells generally grow well, while the number of the bacterium is low and often easily to be neglected. B: the bacterium becomes overwhelming when the growth of host cells is declined, due to a low-density passage, or delayed medium changes. C: bacterium in the early “sessile” stage. See the inset for details. D: bacterium in the swimming stage with 10 μg/mL ciprofloxacin and 10 μg/mL piperacillin added in the medium. See the inset for details. E-F: the RAW 264.7 cells after resuscitation. E: the RAW 264.7 cells after resuscitation without wash; F: only a few RAW 264.7 cells were left after wash. |
When the host cells were passaged at a lower density,or the changes of culture media were delayed,the bacterium would become overwhelming (Figure1-B). Since most of the bacterium swum in the culture,this stage was described as the “swimming” stage. The bacterium was mainly rod-shaped,occurring singly,with short or long chains,and varied from 1.7 to 6.6 μm in length. When piperacillin was included in the medium,it could elongate up to 56 μm long (Figure1-B and C). This seemed to result from incomplete division,since the bacterium was segmented,and looked like a chain consisting of 5-30 connected rod-like structures. It moved much faster with a speed of 19.82-27.87 μm/s and could swim for a long distance. The bacterium collected at this stage was Gram staining negative (Figure2-D). It was also negative for acid-fast staining (Data not shown).
|
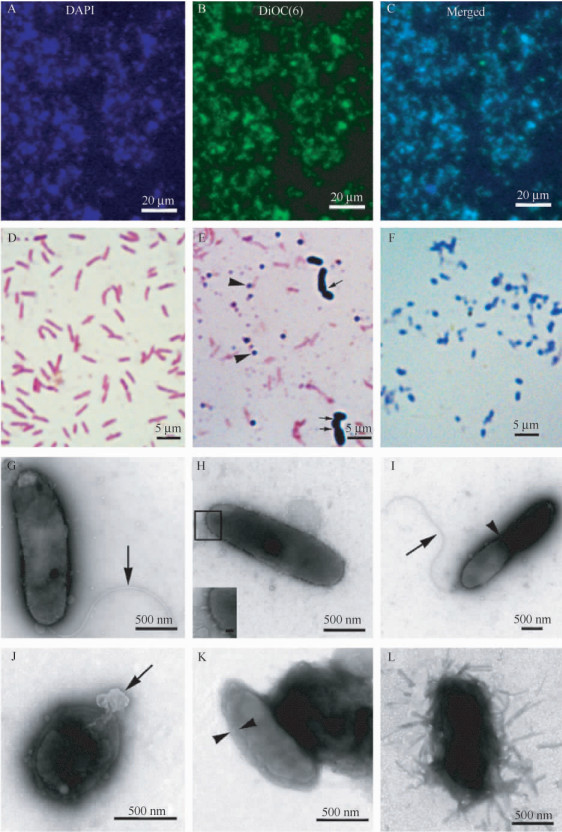
| Figure. 2 Morphological observations of the contaminant. A: DAPI staining; B: DiOC6(3) staining; C: merged from A and B. Scale bars: 20 μm. D-F: gram staining. Scale bars: 5 μm. D: negative staining of the bacterium in the swimming stage; E: both of positive and negative staining of the bacterium can be found after 1 day following the swimming stage. Arrowheads indicate positive rods. Arrows indicate long chained positive structures. F: positive staining in the later sessile stage. G-I: Ultrastructural observations of the bacterium collected in the swimming stage. The inset in H is enlarged in panel H. Scale bars: 500 nm; inset, 50 nm. Arrows in G and I indicate the polar flagellum; Arrowhead in I indicates the binary fission. J-L: Ultrastructural observations of the bacterium collected in the later sessile stage. The arrow in J indicates a protruding process from the bacterium, the nature of which is not known. The arrowheads in K indicate the thickened envelope. In addition, needle-like structures were sometimes present around the envelope (L). Scale bars: 500nm. |
The swimming stage generally lasted less than 1 day. After that,the chained structures became disconnected,and the culture media was full of small rods again (Figure1-D). The host cells stopped growing,with their cytoplasm filled with rod-like granules. Since the bacterium was adherent to each other forming clumps on the floor of the culture vessels or floating in the medium,this stage was described as the “sessile” stage. The bacterium was mainly rod-shaped with (0.4-0.8) μm×(1.0-3.0) μm,occurring singly or in pairs. The rod-shaped bacterium outside the host cells was stained positively for DAPI (Figure2-A),and also for DiOC6(3) (Figure2-B),which is generally used to reveal endoplasmic reticula or mitochondria-like structures in live cells. Different from those in the swimming stage,the bacterium collected at this stage was Gram staining positive (Figure2-E and F).
The contamination with this bacterium frequently led to failed resuscitation of the thawed cells,as shown in a representative image (Figure1-E). Under higher magnification,numerous small rods were seen swimming in the culture,but often indistinguishable from the cell debris. Most of them could be washed off,but some still remained (Figure1-F).
2.2 Electron microscopic observationsUnder the electron microscope,the bacterium collected in the swimming stage appeared rod-shaped (Figure2G-I). Most of them had a polar flagellum (Figure2-G and I),but some without flagellum were also present (Figure2-H). The bacterium was found to reproduce through binary fission (Figure2-I). The bacterium collected in the sessile stage was mainly short rod-like in shape,but flagella were rarely seen (Figure2-J-L). Different from that in the swimming stage,its envelope appeared much thicker (40.90 ±9.38 nm in the sessile stage vs 16.99 ±3.98 nm in the swimming stage, P<0.001,t-test) (Figure2-K),and needle-like structures were sometimes present around the envelope (Figure2-L).
In order to correctly measure their sizes,only the single bacteria were randomly selected. The short diameters ranged from 0.36 to 0.75 μm (Mean ±SD,0.58 ±0.09 μm,n=120),while the long diameter from 1.0 to 2.6 μm (Mean ±SD,1.44 ±0.56 μm,n=120).
2.3 Bacterial culture,sequencing and identificationThe bacterium grew well on LB agar,but was more easily cultured in liquid medium such as DMEM. The bacterial colonies cultured on LB agar for 3 days were round and opaque with neat edges (Figure3-A). Their surfaces were smooth,and shiny with pale yellow. Different from that co-existing with host cells,the bacterium cultured with DMEM was motionless (Figure3-B). Ultrastructural examinations further confirmed that the bacterium collected from the DMEM was non-flagellated. The culture experiments with and without host cells have been repeated more than 3 times,and almost all the bacteria were found attached to the floor of culture vessels.

|
| Figure. 3 Bacterial cultures and antibiotic treatment. Scale bars: A, 2mm; B-F, 20μm. A: the bacterial colonies cultured on LB agar for 3 days are round and opaque with neat edges. The surfaces are smooth, and shiny with pale yellow. B: the bacterium cultured with DMEM is motionless, and attached to the floor of culture vessels. C and D show almost no rod-like structures in RAW 264.7 and SH-SY5Y cultures after antibiotic treatment, respectively. However, the cell surface of SH-SY5Y exhibits numerous spine-like structures. E and F are used as controls, showing a large number of rod-like structures in the RAW 264.7 and SH-SY5Y cultures without antibiotics, respectively. |
The 16S rRNA sequence (GenBank accession number: KC928402) showed 99% identity with that of Phenylobacterium zucineum HLK1T (CP000747). Moreover,sequence analysis further demonstrated that both the Gram staining negative and positive bacteria had the same 16S rRNA gene sequence.
2.4 Antibiotic treatmentsSeveral antibiotics were shown able to inhibit the growth of the bacterium on the LB agar,however,except for imipenem all the antibiotics proved to be ineffective for the bacterium in the liquid media. The inclusion of 400 μg/mL imipenem in the media remarkably inhibited the proliferation of bacteria outside the cells. The macrophage RAW 264.7 cells were also susceptible to the contamination,but if imipenem was continuously added in the medium for 1 week,the bacterium could be successfully removed (Figure3-C) and no bacterium appeared even no drug was added for another week. By contrast,although its number became apparently decreased in the culture media of other cell lines such as SH-SY5Y cells when imipenem was used (Figure3-D),the bacterium appeared again once drug withdrawal,suggesting that imipenem was not effective in removing this microorganism in these cell lines.
2.5 Resuscitation of the thawed cellsThe bacterium frequently became indiscernible in some culture vessels,while the cells looked healthy again. The cells that grew well again were selected and routinely frozen. Meanwhile,the supernatants were also kept as “rescuing” media for resuscitating the cells.
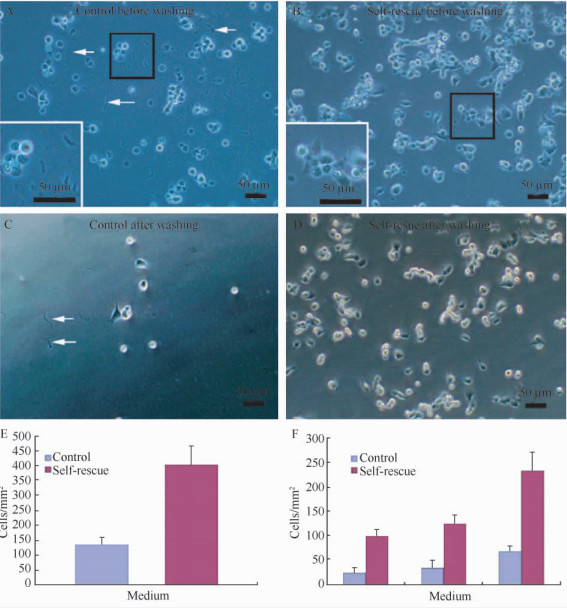
This simple method had remarkably promoted the survival of the thawed cells (Figure4). As compared to those cultured with the freshly prepared media,the cells in the supernatants were increased in number by 3-4 folds (Figure4-C and D). Statistical analysis showed that this resuscitating method was useful for all the cell lines tested (P<0.005 for SH-SY5Y cells,n=6,Student’s t test,Figure4-E; P<0.0001 for RAW 264.7 cells,one-way ANOVA,n=6,Figure4-F). Moreover,the bacterium was generally indiscernible.
|
| Figure. 4 Self-rescuing the contaminated cells.Scale bars: 50 μm. A and B: SH-SY5Y cells in fresh complete medium; C and D: SH-SY5Y cells in self-rescuing culture. The insets in A and C are enlarged in each panel, respectively. E: the number of SH-SY5Y cells (cells/mm2) in self-rescuing medium is significantly larger than that in control medium (P<0.005, Student’s t test, n=6). F: the statistical analysis of self-rescuing RAW 264.7 macrophage cells, which have been counted continuously for 3 days after thawing (P<0.0001, one-way ANOVA, n=6). |
In contrast,many rods appeared again in the cultures using the fresh media,suggesting that the health-looking cells were still the bacterial carrier (Figure4-A and B).
2.6 The infection of SH-SY5Y cell lineBy infecting healthy SH-SY5Y cells with the bacterium of different concentrations,we found that after 24 hours following the infection some cells became dead,and floated in the culture medium. The number of cells was significantly decreased with the increase of the bacterium concentration and the extension of incubation time,as shown in Figure5. In the wells which were added with 20 μL of 108 CFU/mL of the bacterium,there were no survived cells left after 48 hours.

|
| Figure. 5 The number (cells/mm2) of SH-SY5Y cells exposed to different bacterial concentrations at different time (*P<0.005, **P<0.001 vs 0h after infection, t-test, n=6). |
This work was stimulated by the discovery of unknown swimming organisms in the culture supernatants. It was Gram staining negative in the swimming stage,but Gram staining positive in the sessile stage. Electron microscopic examinations revealed that the bacterium collected in the swimming stage had a polar flagellum,and therefore was motile,while that adherent to surfaces or floating in the media in the sessile stage rarely possessed flagellum-like structures. The 16S rRNA sequence analysis showed that this rod-shaped microorganism belonged to the family Caulobacteraceae,class Alphaproteobacteria,and was most closely related to Phenylobacterium zucineum HLK1 T strain with 99% similarity. Because the bacterium from different growth stages showed the same 16S rRNA gene sequence,the presence of Gram staining positive and non-flagellated bacterium seemed not result from an additional contamination by other bacteria,but possibly from the pleomorphism of this kind of bacterium in different habitat conditions[11].
Comparative genomic analysis as well as 16S rRNA gene sequencing demonstrated that Phenylobacterium zucineum HLK1 T is phylogenetically the closest to Caulobacer crescentus[12],which displays a dimorphic life cycle,with a swimming stage and a sessile stalked stage[13, 14]. Only the swimming type is motile with a polar flagellum,but it can differentiate into the sessile type by attaching to each other or to surfaces. Based upon our results,it is very possible for the Phenylobacterium to have a similar dimorphic life cycle. Previous studies showed that Phenylobacterium zucineum HLK1T is motile with a polar flagellum[12, 15],however,neither Gram staining positive nor non-flagellated type has been reported yet. In our case,the contaminated culture was kept at either 37℃ or at room temperature without further medium change for a long time,thus the bacterium had to be confronted to decreased nutrients. By contrast,the bacterium,which was isolated from K562 cells[15],seemed not have been tested under famine conditions. Therefore,the data reported previously seem more pertinent to the swimming type.
The available data suggest that Phenylobacterium is a ubiquitous oligotrophic bacterium found in various habitat conditions,including wastewater from a detergent factory[7],the free-flowing waters of a bore well[6],alkaline groundwater with a pH 11.4[16],activated sludge from waster water treatment plant[17],cotton waste compost[18],fresh water[19] and beach soil[20]. Among them,Phenylobacterium zucineum HLK1T is known as the only Phenylobacterium which can infect the mammalian cells[15]. Our results further showed that the Phenylobacterium can infect all the mammalian cell lines used in this study and that the presence of eukaryotic cells is not necessary. Since it can grow on LB agar,in DMEM with or without host cells,this bacterium seems to belong to environmental bacteria like the other members in the Phenylobacterium,therefore may be present in the environments.
Several members of the family Caulobacteraceae display resistance to alkaline and acidic PH,heat shock,and extreme cold conditions[21]. For example,Caulobacer crescentus has been found to display remarkable survival even frozen at-80℃ for a long time[21]. Genomic analysis showed that Phenylobacterium zucineum HLK1T contains abundant two-component elements,which positively correlate with the capacity for environmental adaptation and for a variety of stresses,including cellular energy depletion,extreme concentrations of various toxic substances[12].
With respect to its strong vitality,it is interesting that this bacterium frequently became indiscernible when co-cultured with the host cells. Since the latter are able to excrete metabolites as well as discharge a variety of diffusible factors into the media to modify their own growth[22, 23],we speculated that some diffusible factors secreted by the cultured cells can inhibit this opportunistic bacterium.
It is a long time that bacterial populations have not been considered as more than the collections of single cells. Until recently,a variety of microorganisms were found capable of secreting autoinducers to minimize the host defense response by modulating their own population density and gene expression[24, 25, 26, 27]. On the other hand,the defense response in higher organisms is generally thought to be fulfilled by professional cells,and it has not been investigated whether the non-professional cells play a similar active role or not.
The contamination with this bacterium frequently led to failed resuscitation of thawed cells. In this study,we found a simple method to resuscitate the thawed cells with the culture supernatants,which remarkably promoted the survival of the thawed cells. Moreover,the number of bacterium was generally decreased dramatically in the used media,the reason for which is not clarified in this study,but we proposed that the culture supernatants may contained some diffusible factors against the bacteria.
| [1] | Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell, 1978, 15:261-267. |
| [2] | Bertram JS, Janik P. Establishment of a cloned line of Lewis lung carcinoma cells adapted to cell culture. Cancer Letters, 1980, 11:63-73. |
| [3] | Klebe RJ, Ruddle FH. Neuroblastoma:Cell culture analysis of a differentiating stem cell system. The Journal of Cell Biology, 1969, 43:69A. |
| [4] | Olmsted JB, Carlson K, Klebe R, Ruddle F, Rosenbaum J. Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proceedings of the National Academy of Sciences of the United States of America, 1970, 65:129-136. |
| [5] | Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. Journal of Structural Biology, 2005, 151:182-195. |
| [6] | Kanso S, Patel BK. Phenylobacterium lituiforme sp. nov., a moderately thermophilic bacterium from a subsurface aquifer, and emended description of the genus Phenylobacterium. International Journal of Systematic and evolutionary Microbiology, 2004, 54:2141-2146. |
| [7] | Ke N, Xiao C, Ying Q, Ji S. A new species of the genus Phenylobacterium for the degradation of LAS (linear alkylbenzene sulfonate). Acta Microbiologica Sinica, 2003, 43(1):1-7.(in Chinese)柯娜,肖昌松,应启锋,纪树兰.一个可降解直链烷基苯磺酸盐的新种.微生物学报,2003, 43(1):1-7. |
| [8] | Zhu XH, Li F, Xu JH, Xiang LH, Kang KF. Cutaneous infectious granuloma caused by Phenylobacterium in an adult with myelodysplastic syndrome:a first case report. American Journal of Clinical Dermatology, 2010, 11:363-366. |
| [9] | Lelong-Rebel IH, Piemont Y, Fabre M, Rebel G. Mycobacterium avium-intracellulare infection of mammalian cell cultures. In vitro Cellular & Developmental Biology. Animal, 2009, 45:75-90. |
| [10] | Gray JS, Birmingham JM, Fenton JI. Got black swimming dots in your cell culture? Identification of Achromobacter as a novel cell culture contaminant. Biologicals, 2010, 38:273-277. |
| [11] | Hirsch P. Microbial life at extremely low nutrient levels. Advances in Space research:the Official Journal of the Committee on Space Research (COSPAR), 1986, 6:287-298. |
| [12] | Luo Y, Xu X, Ding Z, Liu Z, Zhang B, Yan Z, Sun J, Hu S, Hu X. Complete genome of Phenylobacterium zucineum-a novel facultative intracellular bacterium isolated from human erythroleukemia cell line K562. BMC Genomics, 2008, 9:386. |
| [13] | Brun YV, Marczynski G, Shapiro L. The expression of asymmetry during Caulobacter cell differentiation. Annual Review of Biochemistry, 1994, 63:419-450. |
| [14] | Bodenmiller D, Toh E, Brun YV. Development of surface adhesion in Caulobacter crescentus. Journal of Bacteriology, 2004, 186:1438-1437. |
| [15] | Zhang K, Han W, Zhang R, Xu X, Pan Q, Hu X. Phenylobacterium zucineum sp. nov., a facultative intracellular bacterium isolated from a human erythroleukemia cell line K562. Systematic and Applied Microbiology, 2007,30:207-212. |
| [16] | Tiago I, Mendes V, Pires C, Morais PV, Ver Ds' simo A. Phenylobacterium falsum sp. nov., an Alphaproteobacterium isolated from a nonsaline alkaline groundwater, and emended description of the genus Phenylobacterium. Systematic and Applied Microbiology, 2005, 28:295-302. |
| [17] | Aslam Z, Im WT, Ten LN, Lee ST. Phenylobacterium koreense sp. nov., isolated from South Korea. International Journal of Systematic and Evolutionary Microbiology, 2005, 55:2001-2005. |
| [18] | Weon HY, Kim BY, Kwon SW, Go SJ, Koo BS, Stackebrandt E. Phenylobacterium composti sp. nov., isolated from cotton waste compost in Korea. International Journal of Systematic and Evolutionary Microbiology, 2008, 58:2301-2304. |
| [19] | Abraham WR, Macedo AJ, Lünsdorf H, Fischer R, Pawelczyk S, Smit J, Vancanneyt M. Phylogeny by a polyphasic approach of the order Caulobacterales, proposal of Caulobacter mirabilis sp. nov., Phenylobacterium haematophilum sp. nov. and Phenylobacterium conjunctum sp. nov., and emendation of the genus Phenylobacterium. International Journal of Systematic and Evolutionary Microbiology, 2008, 58:1939-1949. |
| [20] | Oh YS, Roh DH. Phenylobacterium muchangponense sp. nov., isolated from beach soil, and emended description of the genus Phenylobacterium. International Journal of Systematic and Evolutionary Microbiology, 2012, 62:977-983. |
| [21] | Mazzon RR, Lang EA, Braz VS, Marques MV. Characterization of Caulobacter crescentus response to low temperature and identification of genes involved in freezing resistance. FEMS Microbiology Letters, 2008,288:178-185. |
| [22] | Lai Y, Sun Y, Skinner CM, Son EL, Lu Z, Tuan RS, Jilka RL, Ling J, Chen XD. Reconstitution of marrow-derived extracellular matrix ex vivo:a robust culture system for expanding large-scale highly functional human mesenchymal stem cells. Stem Cells, 2010, 19:1095-1107. |
| [23] | Sun Y, Li W, Lu Z, Chen R, Ling J, Ran Q, Jilka RL, Chen XD. Rescuing replication and osteogenesis of aged mesenchymal stem cells by exposure to a young extracellular matrix. FASEB Journal:Official Publication of the Federation of American Societies for Experimental Biology, 2011, 25:1474-1485. |
| [24] | Parsek MR, Greenberg EP. Acyl-homoserine lactone quorum sensing in gram-negative bacteria:a signaling mechanism involved in associations with higher organisms. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97:8789-8793. |
| [25] | Reading NC, Sperandio V. Quorum sensing:the many languages of bacteria. FEMS Microbiology Letters, 2006, 254:1-11. |
| [26] | Gomer RH, Jang W, Brazill D. Cell density sensing and size determination. Development, Growth & Differentiation, 2011, 53:482-494. |
| [27] | Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication:acyl-homoserine lactone quorum sensing. Annual Review of Genetics, 2001, 35:439-468. |
 2015, Vol. 55
2015, Vol. 55




